8B9X
 
 | | Chimeric protein of human UFM1 E3 ligase, UFL1, and DDRGK1 | | Descriptor: | CHLORIDE ION, DDRGK domain-containing protein 1,E3 UFM1-protein ligase 1 | | Authors: | Wiener, R, Isupov, M, banerjee, S. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.066 Å) | | Cite: | Structural study of UFL1-UFC1 interaction uncovers the role of UFL1 N-terminal helix in ufmylation.
Embo Rep., 24, 2023
|
|
1A2Z
 
 | |
1UP8
 
 | | Recombinant vanadium-dependent bromoperoxidase from red algae Corallina pilulifera | | Descriptor: | CALCIUM ION, PHOSPHATE ION, VANADIUM-DEPENDENT BROMOPEROXIDASE 1 | | Authors: | Garcia-Rodriguez, E, Isupov, M, Ohshiro, T, Izumi, Y, Littlechild, J.A. | | Deposit date: | 2003-09-29 | | Release date: | 2003-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enhancing Effect of Calcium and Vanadium Ions on Thermal Stability of Bromoperoxidase from Corallina Pilulifera.
J.Biol.Inorg.Chem., 10, 2005
|
|
1QHF
 
 | | YEAST PHOSPHOGLYCERATE MUTASE-3PG COMPLEX STRUCTURE TO 1.7 A | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, PROTEIN (PHOSPHOGLYCERATE MUTASE), SULFATE ION | | Authors: | Crowhurst, G, Littlechild, J, Watson, H.C. | | Deposit date: | 1999-05-13 | | Release date: | 1999-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a phosphoglycerate mutase:3-phosphoglyceric acid complex at 1.7 A.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3LCZ
 
 | | B.licheniformis Anti-TRAP can assemble into two types of dodecameric particles with the same symmetry but inverted orientation of trimers | | Descriptor: | Inhibitor of TRAP, regulated by T-BOX (Trp) sequence RtpA, ZINC ION | | Authors: | Shevtsov, M.B, Chen, Y, Gollnick, P, Antson, A.A. | | Deposit date: | 2010-01-12 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Bacillus licheniformis Anti-TRAP can assemble into two types of dodecameric particles with the same symmetry but inverted orientation of trimers.
J.Struct.Biol., 170, 2010
|
|
4BRZ
 
 | | Haloalkane dehalogenase | | Descriptor: | CHLORIDE ION, HALOALKANE DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2013-06-06 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and Structural Characterisation of a Haloalkane Dehalogenase from a Marine Rhodobacteraceae.
FEBS Lett., 588, 2014
|
|
4B8N
 
 | | Cytochrome b5 of Ostreococcus tauri virus 2 | | Descriptor: | CYTOCHROME B5-HOST ORIGIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Isupov, M, Reid, E.L, Weynberg, K.D, Love, J, Wilson, W.H, Kelly, S.L, Lamb, D.C, Allen, M.J, Littlechild, J.A. | | Deposit date: | 2012-08-28 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional and Structural Characterisation of a Viral Cytochrome B5.
FEBS Lett., 587, 2013
|
|
4C3T
 
 | | The Carbonic anhydrase from Thermovibrio ammonificans reveals an interesting intermolecular disulfide contributing to increasing thermal stability of this enzyme | | Descriptor: | CARBONATE DEHYDRATASE, CHLORIDE ION, ZINC ION | | Authors: | James, P, Isupov, M, Sayer, C, Berg, S, Lioliou, M, Kotlar, H, Littlechild, J. | | Deposit date: | 2013-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The Structure of a Tetrameric [Alpha]-Carbonic Anhydrase from Thermovibrio Ammonificans Reveals a Core Formed Around Intermolecular Disulfides that Contribute to its Thermostability
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4C6H
 
 | | Haloalkane dehalogenase with 1-hexanol | | Descriptor: | CHLORIDE ION, HALOALKANE DEHALOGENASE, HEXAN-1-OL, ... | | Authors: | Novak, H.R, Sayer, C, Isupov, M, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Biochemical and Structural Characterisation of a Haloalkane Dehalogenase from a Marine Rhodobacteraceae.
FEBS Lett., 588, 2014
|
|
4CMD
 
 | | The (R)-selective transaminase from Nectria haematococca | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sayer, C, Isupov, M, Martinez-Torres, R.J, Richter, N, Hailes, H.C, Ward, J, Littlechild, J. | | Deposit date: | 2014-01-16 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Substrate Specificity, Enantioselectivity and Structure of the (R)-Selective Amine:Pyruvate Transaminase from Nectria Haematococca.
FEBS J., 281, 2014
|
|
4CMF
 
 | | The (R)-selective transaminase from Nectria haematococca with inhibitor bound | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, AMINOTRANSFERASE, ... | | Authors: | Sayer, C, Isupov, M, Martinez-Torres, R.J, Richter, N, Hailes, H.C, Ward, J, Littlechild, J. | | Deposit date: | 2014-01-16 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Substrate Specificity, Enantioselectivity and Structure of the (R)-Selective Amine:Pyruvate Transaminase from Nectria Haematococca.
FEBS J., 281, 2014
|
|
6G1C
 
 | |
6G1N
 
 | |
7ZTH
 
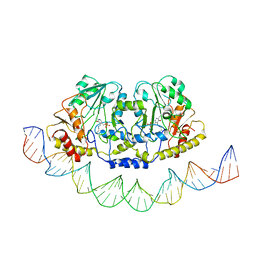 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-05-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZN5
 
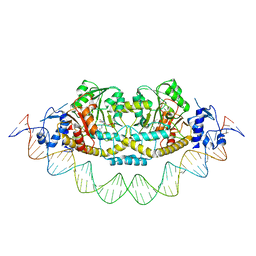 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry. | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZPA
 
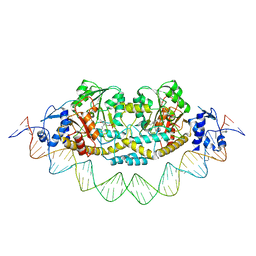 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZLA
 
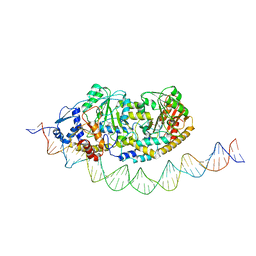 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Savino, C, Exertier, C, Bolognesi, M, Chaves Sanjuan, A. | | Deposit date: | 2022-04-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
8PKB
 
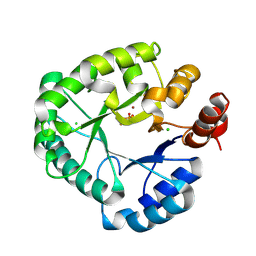 | |
6J4K
 
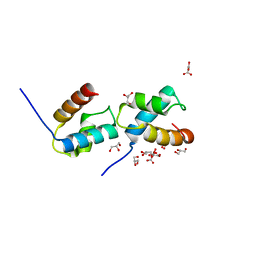 | |
5W19
 
 | | Tryptophan indole-lyase complex with oxindolyl-L-alanine | | Descriptor: | 1-carboxy-1-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]azaniumyl}-2-[(3R)-2-oxo-2,3-dihydro-1H-indol-3-yl]ethan-1-ide, POTASSIUM ION, Tryptophanase | | Authors: | Phillips, R.S, Wood, Z.A. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of Proteus vulgaris tryptophan indole-lyase complexed with oxindolyl-L-alanine: implications for the reaction mechanism.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1QEX
 
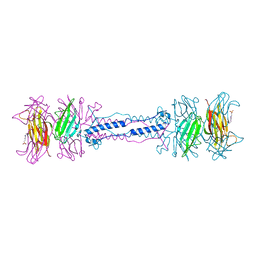 | | BACTERIOPHAGE T4 GENE PRODUCT 9 (GP9), THE TRIGGER OF TAIL CONTRACTION AND THE LONG TAIL FIBERS CONNECTOR | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PROTEIN (BACTERIOPHAGE T4 GENE PRODUCT 9 (GP9)) | | Authors: | Kostyuchenko, V.A, Navruzbekov, G.A, Kurochkina, L.P, Strelkov, S.V, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 1999-03-30 | | Release date: | 1999-10-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of bacteriophage T4 gene product 9: the trigger for tail contraction.
Structure Fold.Des., 7, 1999
|
|
1TPL
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF TYROSINE PHENOL-LYASE | | Descriptor: | SULFATE ION, TYROSINE PHENOL-LYASE | | Authors: | Antson, A, Demidkina, T, Dauter, Z, Harutyunyan, E, Wilson, K. | | Deposit date: | 1992-11-25 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of tyrosine phenol-lyase.
Biochemistry, 32, 1993
|
|
