7MBS
 
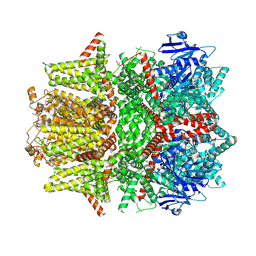 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 6 uM calcium (open state) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBT
 
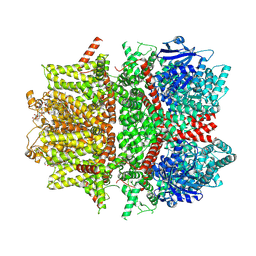 | | Cryo-EM structure of zebrafish TRPM5 E337A mutant in the presence of 5 mM calcium (low calcium occupancy in the transmembrane domain) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBU
 
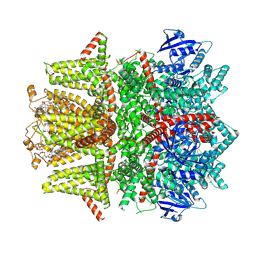 | | Cryo-EM structure of zebrafish TRPM5 E337A mutant in the presence of 5 mM calcium (high calcium occupancy in the transmembrane domain) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBQ
 
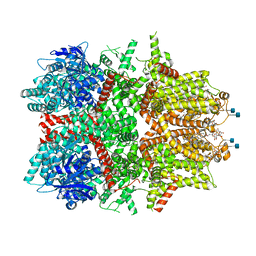 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 5 mM calcium | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBV
 
 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 5 mM calcium and 0.5 mM NDNA | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBP
 
 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 1 mM EDTA | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-22 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5ZNR
 
 | | Crystal structure of PtSHL in complex with an H3K27me3 peptide | | Descriptor: | 17-mer peptide from Histone H3.2, SHORT LIFE family protein, SULFATE ION, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Dual recognition of H3K4me3 and H3K27me3 by a plant histone reader SHL.
Nat Commun, 9, 2018
|
|
6A5M
 
 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 2 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A5N
 
 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with methylated DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*TP*GP*CP*TP*GP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*TP*AP*CP*TP*(5CM)P*AP*GP*CP*AP*GP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A5K
 
 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 1 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZNP
 
 | | Crystal structure of PtSHL in complex with an H3K4me3 peptide | | Descriptor: | 15-mer peptide from Histone H3.2, SHORT LIFE family protein, ZINC ION | | Authors: | Lv, X, Du, J. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual recognition of H3K4me3 and H3K27me3 by a plant histone reader SHL.
Nat Commun, 9, 2018
|
|
7VG2
 
 | | Cryo-EM structure of Arabidopsis DCL3 in complex with a 40-bp RNA | | Descriptor: | CALCIUM ION, Dicer-like 3, TAS1a forward strand (5'-phosphorylation), ... | | Authors: | Wang, Q, Du, J. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation.
Science, 374, 2021
|
|
7VG3
 
 | | Cryo-EM structure of Arabidopsis DCL3 in complex with a 30-bp RNA | | Descriptor: | CALCIUM ION, Dicer-like 3, TAS1a RNA forward strand (5'-phosphorylated), ... | | Authors: | Wang, Q, Du, J. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation.
Science, 374, 2021
|
|
7VSQ
 
 | |
7VSP
 
 | | Crystal strcuture of the tandem B-Box domains of COL2 | | Descriptor: | ZINC ION, Zinc finger protein CONSTANS-LIKE 2 | | Authors: | Lv, X, Liu, R, Du, J. | | Deposit date: | 2021-10-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of CONSTANS oligomerization in FLOWERING LOCUS T activation.
J Integr Plant Biol, 64, 2022
|
|
7CCE
 
 | |
6IP0
 
 | |
6IP4
 
 | |
6J9A
 
 | | Crystal structure of Arabidopsis thaliana VAL1 in complex with FLC DNA fragment | | Descriptor: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*TP*C)-3'), DNA (5'-D(*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*TP*G)-3') | | Authors: | Hu, H, Du, J. | | Deposit date: | 2019-01-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.915 Å) | | Cite: | Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis.
Nat.Plants, 5, 2019
|
|
6J9C
 
 | | Crystal structure of Arabidopsis thaliana transcription factor LEC2-DNA complex | | Descriptor: | B3 domain-containing transcription factor LEC2, DNA (5'-D(*CP*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*T)-3'), DNA (5'-D(*GP*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*T)-3') | | Authors: | Hu, H, Du, J. | | Deposit date: | 2019-01-22 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis.
Nat.Plants, 5, 2019
|
|
6J9B
 
 | | Arabidopsis FUS3-DNA complex | | Descriptor: | B3 domain-containing transcription factor FUS3, DNA (5'-D(*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*TP*C)-3'), DNA (5'-D(*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*TP*G)-3') | | Authors: | Hu, H, Du, J. | | Deposit date: | 2019-01-22 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis.
Nat.Plants, 5, 2019
|
|
7CVQ
 
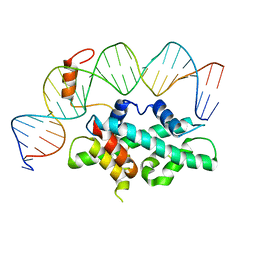 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB2/YC3 and FT CORE1 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-3 and Zinc finger protein CONSTANS, FT CORE1 DNA forward strand, FT CORE1 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
7CVO
 
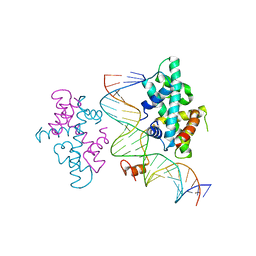 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB3/YC4 and FT CORE2 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-4 and Zinc finger protein CONSTANS, FT CORE2 DNA forward strand, FT CORE2 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
7DE9
 
 | |
7ET5
 
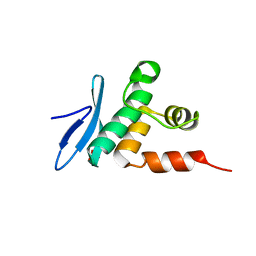 | | Crystal structure of Arabidopsis TEM1 AP2 domain | | Descriptor: | AP2/ERF and B3 domain-containing transcription repressor TEM1, SULFATE ION | | Authors: | Hu, H, Du, J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.052 Å) | | Cite: | TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
