1WP8
 
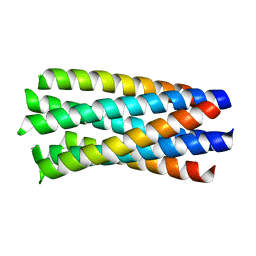 | | crystal structure of Hendra Virus fusion core | | Descriptor: | Fusion glycoprotein F0,Fusion glycoprotein F0 | | Authors: | Xu, Y, Liu, Y, Lou, Z, Su, N, Bai, Z, Gao, G.F, Rao, Z. | | Deposit date: | 2004-08-31 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Nipah and Hendra virus fusion core proteins
FEBS J., 273, 2006
|
|
1X0V
 
 | |
1X0X
 
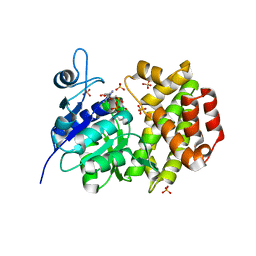 | |
2GER
 
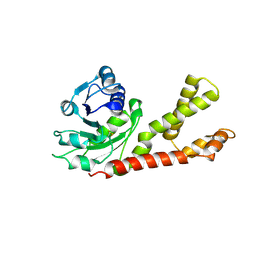 | |
2GR9
 
 | | Crystal structure of P5CR complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLUTAMIC ACID, Pyrroline-5-carboxylate reductase 1 | | Authors: | Meng, Z, Lou, Z, Liu, Z, Rao, Z. | | Deposit date: | 2006-04-23 | | Release date: | 2006-10-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of human pyrroline-5-carboxylate reductase
J.Mol.Biol., 359, 2006
|
|
3CM8
 
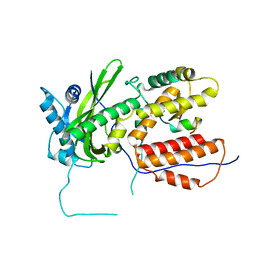 | | A RNA polymerase subunit structure from virus | | Descriptor: | Polymerase acidic protein, peptide from RNA-directed RNA polymerase catalytic subunit | | Authors: | He, X, Zhou, J, Zeng, Z, Ma, J, Zhang, R, Rao, Z, Liu, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2008-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Crystal structure of the polymerase PAC-PB1N complex from an avian influenza H5N1 virus
Nature, 454, 2008
|
|
2GRA
 
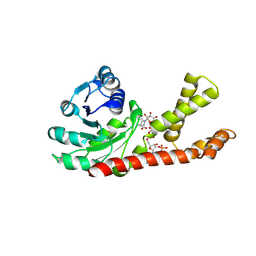 | | crystal structure of Human Pyrroline-5-carboxylate Reductase complexed with nadp | | Descriptor: | GLUTAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pyrroline-5-carboxylate reductase 1 | | Authors: | Meng, Z, Lou, Z, Liu, Z, Rao, Z. | | Deposit date: | 2006-04-23 | | Release date: | 2006-10-03 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of human pyrroline-5-carboxylate reductase
J.Mol.Biol., 359, 2006
|
|
2ZGJ
 
 | | Crystal Structure of D86N-GzmM Complexed with Its Optimal Synthesized Substrate | | Descriptor: | Granzyme M, SSGKVPLS, SULFATE ION | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-01-22 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
3KO1
 
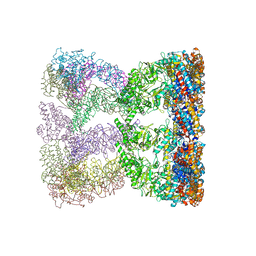 | | Cystal structure of thermosome from Acidianus tengchongensis strain S5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin | | Authors: | Huo, Y, Zhang, K, Hu, Z, Wang, L, Zhai, Y, Zhou, Q, Lander, G, He, Y, Zhu, J, Xu, W, Dong, Z, Sun, F. | | Deposit date: | 2009-11-12 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of group II chaperonin in the open state.
Structure, 18, 2010
|
|
3N6N
 
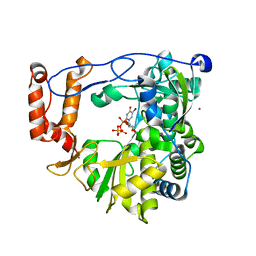 | | crystal structure of EV71 RdRp in complex with Br-UTP | | Descriptor: | 5-bromouridine 5'-(tetrahydrogen triphosphate), NICKEL (II) ION, RNA-dependent RNA polymerase | | Authors: | Wu, Y, Lou, Z.Y, Miao, Y, Yu, Y, Rao, Z.H. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China.
Protein Cell, 1, 2010
|
|
3MTV
 
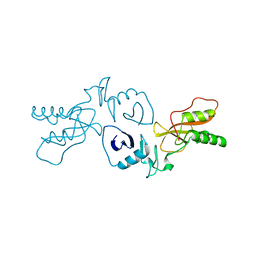 | | The Crystal Structure of the PRRSV Nonstructural Protein Nsp1 | | Descriptor: | Papain-like cysteine protease | | Authors: | Xue, F, Sun, Y.N, Yan, L.M, Zhao, C, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2010-04-30 | | Release date: | 2010-05-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of porcine reproductive and respiratory syndrome virus nonstructural protein Nsp1beta reveals a novel metal-dependent nuclease
J.Virol., 84, 2010
|
|
3N6M
 
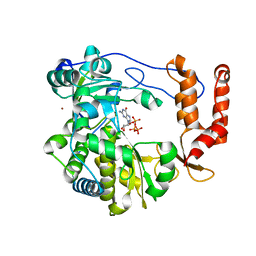 | | Crystal structure of EV71 RdRp in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, NICKEL (II) ION, RNA-dependent RNA polymerase | | Authors: | Wu, Y, Lou, Z.Y, Miao, Y, Yu, Y, Rao, Z.H. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China.
Protein Cell, 1, 2010
|
|
3N6L
 
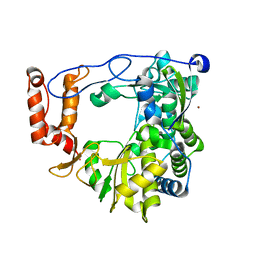 | | The crystal structure of RNA-dependent RNA polymerase of EV71 virus | | Descriptor: | NICKEL (II) ION, RNA-dependent RNA polymerase | | Authors: | Wu, Y, Rao, Z.H. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China.
Protein Cell, 1, 2010
|
|
5F22
 
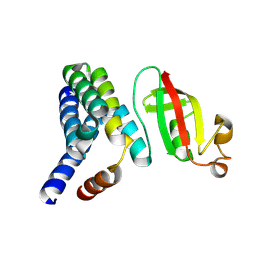 | | C-terminal domain of SARS-CoV nsp8 complex with nsp7 | | Descriptor: | Non-structural protein, Non-structural protein 7 | | Authors: | Li, S. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | C-terminal domain of SARS-CoV nsp8 complex with nsp7
To Be Published
|
|
2FR7
 
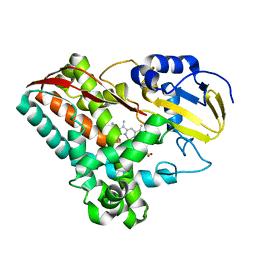 | | Crystal Structure of Cytochrome P450 CYP199A2 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, putative cytochrome P450 | | Authors: | Rao, Z, Wong, L.L, Xu, F, Bell, S.G. | | Deposit date: | 2006-01-19 | | Release date: | 2007-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of CYP199A2, a para-substituted benzoic acid oxidizing cytochrome P450 from Rhodopseudomonas palustris
J.Mol.Biol., 383, 2008
|
|
2ESW
 
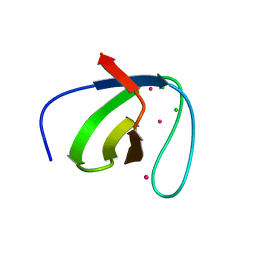 | |
2GA6
 
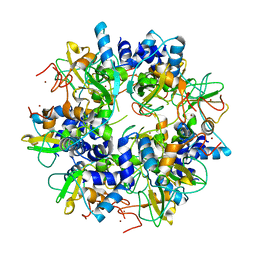 | | The crystal structure of SARS nsp10 without zinc ion as additive | | Descriptor: | ZINC ION, orf1a polyprotein | | Authors: | Su, D, Lou, Z, Sun, F, Zhai, Y, Yang, H, Rao, Z. | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dodecamer Structure of Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein nsp10
J.Virol., 80, 2006
|
|
2GTH
 
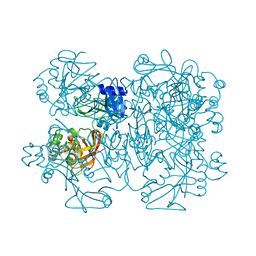 | | crystal structure of the wildtype MHV coronavirus non-structural protein nsp15 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Xu, X, Zhai, Y, Sun, F, Lou, Z, Su, D, Rao, Z. | | Deposit date: | 2006-04-28 | | Release date: | 2006-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | New Antiviral Target Revealed by the Hexameric Structure of Mouse Hepatitis Virus Nonstructural Protein nsp15
J.Virol., 80, 2006
|
|
2G9T
 
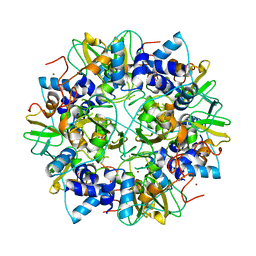 | | Crystal structure of the SARS coronavirus nsp10 at 2.1A | | Descriptor: | ZINC ION, orf1a polyprotein | | Authors: | Su, D, Lou, Z, Yang, H, Sun, F, Rao, Z. | | Deposit date: | 2006-03-07 | | Release date: | 2006-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dodecamer Structure of Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein nsp10
J.Virol., 80, 2006
|
|
2H2Z
 
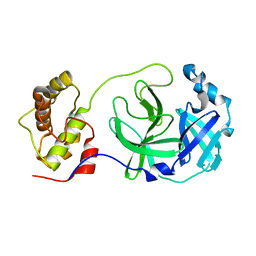 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Yang, H, Xue, X, Shen, W, Zhao, Q, Rao, Z. | | Deposit date: | 2006-05-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
2GTI
 
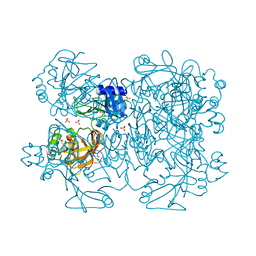 | | mutation of MHV coronavirus non-structural protein nsp15 (F307L) | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab, SULFATE ION | | Authors: | Xu, X, Zhai, Y, Sun, F, Lou, Z, Su, D, Rao, Z. | | Deposit date: | 2006-04-28 | | Release date: | 2006-08-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | New Antiviral Target Revealed by the Hexameric Structure of Mouse Hepatitis Virus Nonstructural Protein nsp15
J.Virol., 80, 2006
|
|
2HOB
 
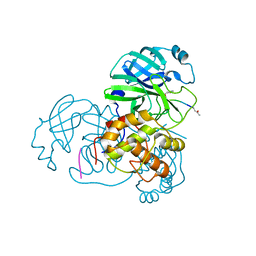 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini in complex with a Michael acceptor N3 | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, Replicase polyprotein 1ab | | Authors: | Xue, X, Yang, H, Shen, W, Zhao, Q, Li, J, Rao, Z. | | Deposit date: | 2006-07-14 | | Release date: | 2007-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
6MYU
 
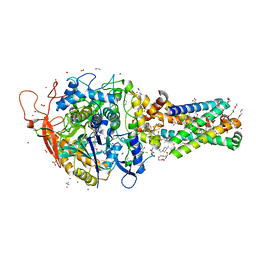 | | Avian mitochondrial complex II crystallized in the presence of HQNO | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6MYT
 
 | | Avian mitochondrial complex II with Atpenin A5 bound, sidechain in pocket | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 3-[(2S,4S,5R)-5,6-DICHLORO-2,4-DIMETHYL-1-OXOHEXYL]-4-HYDROXY-5,6-DIMETHOXY-2(1H)-PYRIDINONE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6MYR
 
 | | Avian mitochondrial complex II with thiapronil bound | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Huang, L.-S, Luemmen, P, Berry, E.A. | | Deposit date: | 2018-11-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
