2FSY
 
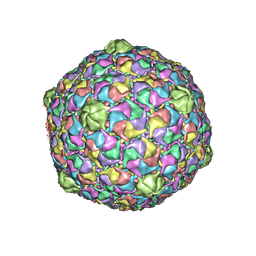 | | Bacteriophage HK97 Pepsin-treated Expansion Intermediate IV | | Descriptor: | major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-23 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
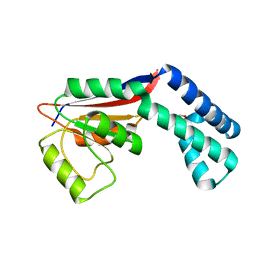
2FZL
 
 | |
2FWR
 
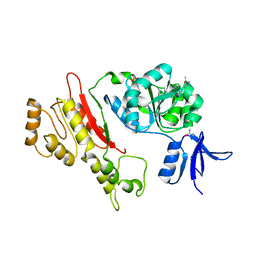 | | Structure of Archaeoglobus Fulgidis XPB | | Descriptor: | DNA repair protein RAD25, ISOPROPYL ALCOHOL, PHOSPHATE ION | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2006-02-02 | | Release date: | 2006-04-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conserved XPB Core Structure and Motifs for DNA Unwinding: Implications for Pathway Selection of Transcription or Excision Repair
Mol.Cell, 22, 2006
|
|
2FRP
 
 | | Bacteriophage HK97 Expansion Intermediate IV | | Descriptor: | Major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-19 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (7.5 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
6YX9
 
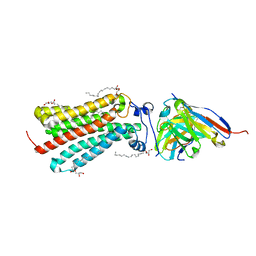 | | Cryogenic human adiponectin receptor 2 (ADIPOR2) at 2.4 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, GLYCEROL, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXG
 
 | | Cryogenic human adiponectin receptor 2 (ADIPOR2) with Tb-XO4 ligand determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, OLEIC ACID, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXD
 
 | | Room temperature structure of human adiponectin receptor 2 (ADIPOR2) at 2.9 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, GLYCEROL, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXF
 
 | | Cryogenic human adiponectin receptor 2 (ADIPOR2) with Gd-DO3 ligand determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, Adiponectin receptor protein 2, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.025 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXH
 
 | | Cryogenic human alkaline ceramidase 3 (ACER3) at 2.6 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | Alkaline ceramidase 3, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
2FT1
 
 | | Bacteriophage HK97 Head II | | Descriptor: | major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-23 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
2FS2
 
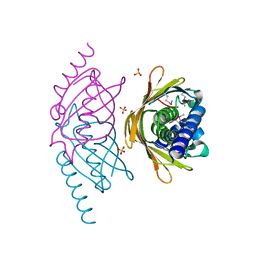 | | Structure of the E. coli PaaI protein from the phyenylacetic acid degradation operon | | Descriptor: | Phenylacetic acid degradation protein paaI, SULFATE ION | | Authors: | Kniewel, R, Buglino, J.A, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-01-20 | | Release date: | 2006-02-07 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, Function, and Mechanism of the Phenylacetate Pathway Hot Dog-fold Thioesterase PaaI
J.Biol.Chem., 281, 2006
|
|
2FZU
 
 | | Reduced enolate chromophore intermediate for GFP variant | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2006-02-10 | | Release date: | 2006-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural evidence for an enolate intermediate in GFP fluorophore biosynthesis.
J.Am.Chem.Soc., 128, 2006
|
|
2FS3
 
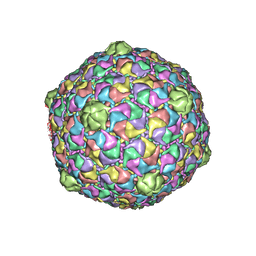 | | Bacteriophage HK97 K169Y Head I | | Descriptor: | Major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-20 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
6ZCU
 
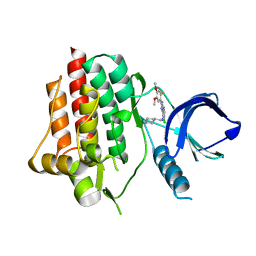 | | syk in complex with 57262_SYKB-AZ13344324-2 | | Descriptor: | 6-chloro-2-(2-fluoro-4,5-dimethoxyphenyl)-N-(piperidin-4-ylmethyl)-3H-imidazo[4,5-b]pyridin-7-amine, Tyrosine-protein kinase SYK | | Authors: | Read, J.A, Patel, J. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 2b
To Be Published
|
|
6ZCP
 
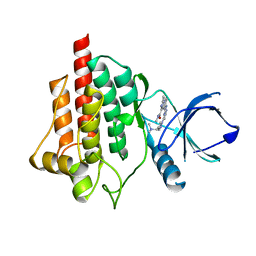 | | SYK Kinase domain in complex with azabenzimidazole inhibitor 2b | | Descriptor: | 7-[[4-[4-[(dimethylamino)methyl]pyrazol-1-yl]pyrimidin-2-yl]amino]-~{N},3-dimethyl-imidazo[1,5-a]pyridine-1-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Read, J.A. | | Deposit date: | 2020-06-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 4
To Be Published
|
|
6ZCX
 
 | | SYK Kinase domain in complex with azabenzimidazole inhibitor 18 | | Descriptor: | Tyrosine-protein kinase SYK, ~{N},1-dimethyl-5-[[4-[3-methyl-4-[[(3~{R})-piperidin-3-yl]amino]pyrazol-1-yl]pyrimidin-2-yl]amino]indazole-3-carboxamide | | Authors: | Read, J.A. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 18
To Be Published
|
|
6ZCQ
 
 | | SYK Kinase domain in complex with diamine inhibitor 5 | | Descriptor: | 7-[[4-[4-[[(3~{R},4~{R})-3-azanyloxan-4-yl]amino]pyrazol-1-yl]pyrimidin-2-yl]amino]-~{N},3-dimethyl-imidazo[1,5-a]pyridine-1-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Read, J.A. | | Deposit date: | 2020-06-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | SYK Kinase domain in complex with diamine inhibitor 5
To Be Published
|
|
6ZCS
 
 | | SYK Kinase domain in complex with azabenzimidazole inhibitor 3 | | Descriptor: | (3~{R},4~{R})-~{N}4-[1-[2-(1-methylindol-4-yl)-3~{H}-imidazo[4,5-b]pyridin-7-yl]pyrazol-4-yl]oxane-3,4-diamine, Tyrosine-protein kinase SYK | | Authors: | Read, J.A. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 3
To Be Published
|
|
6ZCR
 
 | | SYK Kinase domain in complex with azabenzimidazole inhibitor 7 | | Descriptor: | 6-chloro-2-(2-fluoro-4,5-dimethoxyphenyl)-N-(piperidin-4-ylmethyl)-3H-imidazo[4,5-b]pyridin-7-amine, Tyrosine-protein kinase SYK | | Authors: | Read, J.A, Patel, J. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 7
To Be Published
|
|
6ZC0
 
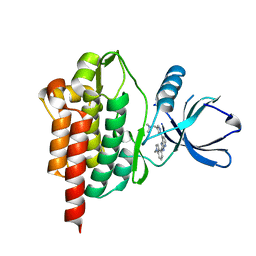 | | SYK Kinase domain in complex with azabenzimidazole inhibitor 2b | | Descriptor: | Tyrosine-protein kinase SYK, ~{N},~{N}-dimethyl-1-[1-[2-(1-methylindol-4-yl)-3~{H}-imidazo[4,5-b]pyridin-7-yl]pyrazol-4-yl]methanamine | | Authors: | Read, J.A. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 2b
To Be Published
|
|
6ZCY
 
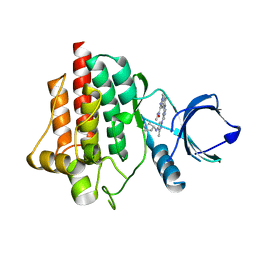 | | SYK Kinase domain in complex with diamine inhibitor 16 | | Descriptor: | Tyrosine-protein kinase SYK, ~{N},1-dimethyl-5-[[4-[3-methyl-4-[[(3~{R})-pyrrolidin-3-yl]amino]pyrazol-1-yl]pyrimidin-2-yl]amino]indazole-3-carboxamide | | Authors: | Read, J.A. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 16
To Be Published
|
|
2FTE
 
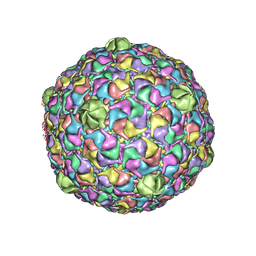 | | Bacteriophage HK97 Expansion Intermediate IV | | Descriptor: | major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-24 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
6ZSL
 
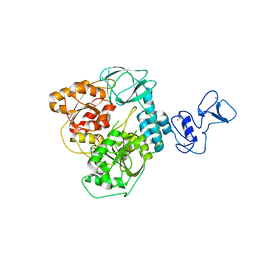 | | Crystal structure of the SARS-CoV-2 helicase at 1.94 Angstrom resolution | | Descriptor: | PHOSPHATE ION, SARS-CoV-2 helicase NSP13, ZINC ION | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Arrowsmith, C.H, von Delft, F, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-07-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
2G4C
 
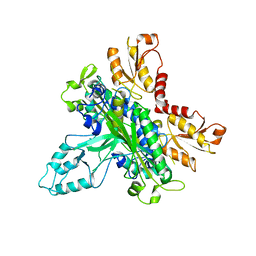 | | Crystal Structure of human DNA polymerase gamma accessory subunit | | Descriptor: | DNA polymerase gamma subunit 2 | | Authors: | Fan, L, Farr, C.L, Kaguni, L.S, Tainer, J.A. | | Deposit date: | 2006-02-22 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A novel processive mechanism for DNA synthesis revealed by structure, modeling and mutagenesis of the accessory subunit of human mitochondrial DNA polymerase
J.Mol.Biol., 358, 2006
|
|
2GD4
 
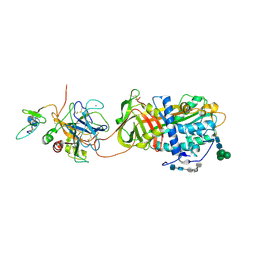 | | Crystal Structure of the Antithrombin-S195A Factor Xa-Pentasaccharide Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, ... | | Authors: | Johnson, D.J, Li, W, Adams, T.E, Huntington, J.A. | | Deposit date: | 2006-03-15 | | Release date: | 2006-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Antithrombin-S195A factor Xa-heparin structure reveals the allosteric mechanism of antithrombin activation.
Embo J., 25, 2006
|
|
