2IG2
 
 | | DIR PRIMAERSTRUKTUR DES KRISTALLISIERBAREN MONOKLONALEN IMMUNOGLOBULINS IGG1 KOL. II. AMINOSAEURESEQUENZ DER L-KETTE, LAMBDA-TYP, SUBGRUPPE I (GERMAN) | | Descriptor: | IGG1-LAMBDA KOL FAB (HEAVY CHAIN), IGG1-LAMBDA KOL FAB (LIGHT CHAIN) | | Authors: | Marquart, M, Huber, R. | | Deposit date: | 1989-04-18 | | Release date: | 1989-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The primary structure of crystallizable monoclonal immunoglobulin IgG1 Kol. II. Amino acid sequence of the L-chain, gamma-type, subgroup I
Biol.Chem.Hoppe-Seyler, 370, 1989
|
|
2IDQ
 
 | | Structure of M98A mutant of amicyanin, Cu(II) | | Descriptor: | Amicyanin, COPPER (II) ION, PHOSPHATE ION | | Authors: | Carrell, C.J, Ma, J.K, Antholine, W, Hosler, J.P, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-09-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Generation of Novel Copper Sites by Mutation of the Axial Ligand of Amicyanin. Atomic Resolution Structures and Spectroscopic Properties
Biochemistry, 46, 2007
|
|
4UT6
 
 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE2 B7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 B7, ... | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4UTA
 
 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE1 C8 | | Descriptor: | BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8 HEAVY CHAIN, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8 LIGHT CHAIN, ENVELOPE GLYCOPROTEIN E, ... | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
5YA2
 
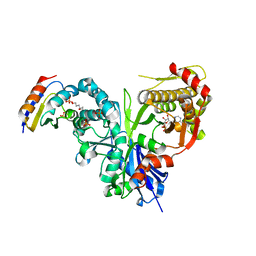 | | Crystal structure of LsrK-HPr complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Autoinducer-2 kinase, HEXANE-1,6-DIOL, ... | | Authors: | Ryu, K.S, Ha, J.H. | | Deposit date: | 2017-08-30 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Evidence of link between quorum sensing and sugar metabolism inEscherichia colirevealed via cocrystal structures of LsrK and HPr
Sci Adv, 4, 2018
|
|
5YA1
 
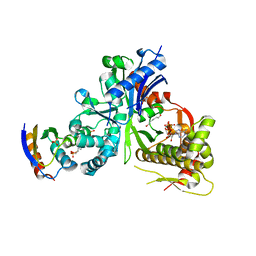 | | crystal structure of LsrK-HPr complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Autoinducer-2 kinase, HEXANE-1,6-DIOL, ... | | Authors: | Ryu, K.S, Ha, J.H. | | Deposit date: | 2017-08-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Evidence of link between quorum sensing and sugar metabolism inEscherichia colirevealed via cocrystal structures of LsrK and HPr
Sci Adv, 4, 2018
|
|
5XYV
 
 | |
5YA0
 
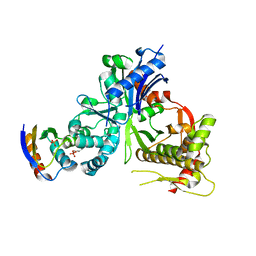 | | Crystal structure of LsrK and HPr complex | | Descriptor: | Autoinducer-2 kinase, HEXANE-1,6-DIOL, PHOSPHATE ION, ... | | Authors: | Ryu, K.S, Ha, J.H. | | Deposit date: | 2017-08-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Evidence of link between quorum sensing and sugar metabolism inEscherichia colirevealed via cocrystal structures of LsrK and HPr
Sci Adv, 4, 2018
|
|
1CLL
 
 | |
1CAR
 
 | | I-CARRAGEENAN. MOLECULAR STRUCTURE AND PACKING OF POLYSACCHARIDE DOUBLE HELICES IN ORIENTED FIBRES OF DIVALENT CATION SALTS | | Descriptor: | 4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose | | Authors: | Arnott, S. | | Deposit date: | 1978-05-23 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-07 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | Iota-carrageenan: molecular structure and packing of polysaccharide double helices in oriented fibres of divalent cation salts.
J.Mol.Biol., 90, 1974
|
|
8DTK
 
 | |
8EUS
 
 | | Crystal structure of NPC1 luminal domain C | | Descriptor: | NPC intracellular cholesterol transporter 1 | | Authors: | Odongo, L, Pornillos, O. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Purification and structure of luminal domain C of human Niemann-Pick C1 protein.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
9GV5
 
 | |
5IIE
 
 | |
9FOT
 
 | | Human monocarboxylate transporter 8 bound to Silychristin | | Descriptor: | (2~{R},3~{R})-2-[(2~{S},3~{R})-3-(hydroxymethyl)-2-(3-methoxy-4-oxidanyl-phenyl)-7-oxidanyl-2,3-dihydro-1-benzofuran-5-yl]-3,5,7-tris(oxidanyl)-2,3-dihydrochromen-4-one, Alfa-tag binding nanobody, Monocarboxylate transporter 8 | | Authors: | Coscia, F, Tassinari, M. | | Deposit date: | 2024-06-12 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular mechanism of thyroxine transport by monocarboxylate transporters.
Nat Commun, 16, 2025
|
|
9GF8
 
 | | Human Monocarboxylate Transporter 8 | | Descriptor: | ALFA-tag binding nanobody, Monocarboxylate transporter 8 | | Authors: | Coscia, F, Tassinari, M. | | Deposit date: | 2024-08-08 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular mechanism of thyroxine transport by monocarboxylate transporters.
Nat Commun, 16, 2025
|
|
9FKN
 
 | | Human monocarboxylate transporter 8 bound to thyroxine | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, ALFA-tag nanobody, Monocarboxylate transporter 8 | | Authors: | Coscia, F, Tassinari, M. | | Deposit date: | 2024-06-03 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular mechanism of thyroxine transport by monocarboxylate transporters.
Nat Commun, 16, 2025
|
|
9GSZ
 
 | |
8EM8
 
 | | Co-crystal structure of the cGMP-dependent protein kinase PKG from Plasmodium falciparum in complex with RY-1-165 | | Descriptor: | UNKNOWN ATOM OR ION, [(3R)-3-{[(4M)-4-(4-cyclopropyl-2-phenyl-1H-imidazol-1-yl)pyrimidin-2-yl]amino}pyrrolidin-1-yl](1,3-thiazol-2-yl)methanone, cGMP-dependent protein kinase, ... | | Authors: | Hutchinson, A, Dong, A, Seitova, A, Bhanot, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-09-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure-Activity Relationship of a Pyrrole Based Series of PfPKG Inhibitors as Anti-Malarials.
J.Med.Chem., 67, 2024
|
|
8FO0
 
 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to a partially cleaved SAM molecule | | Descriptor: | IRON/SULFUR CLUSTER, POTASSIUM ION, Pyruvate formate-lyase 1-activating enzyme, ... | | Authors: | Moody, J.D, Saxton, A.J, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2022-12-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
8FOL
 
 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to SAM, alternate crystal form | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | Authors: | Moody, J.D, Saxton, A.J, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2022-12-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
5J3C
 
 | |
5J30
 
 | |
5CZP
 
 | | 70S termination complex containing E. coli RF2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Dunham, C.M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-10-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.29995918 Å) | | Cite: | Uniformity of Peptide Release Is Maintained by Methylation of Release Factors.
Cell Rep, 17, 2016
|
|
4LZ5
 
 | |
