3LNQ
 
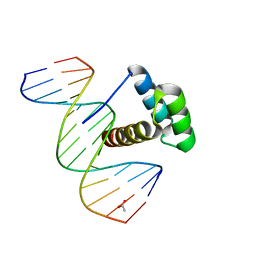 | | Structure of Aristaless homeodomain in complex with DNA | | Descriptor: | 5'-D(*CP*CP*CP*TP*AP*AP*TP*TP*AP*AP*AP*CP*CP*C)-3', 5'-D(*GP*GP*GP*TP*TP*TP*AP*AP*TP*TP*AP*GP*GP*G)-3', ACETATE ION, ... | | Authors: | Takamura, Y, Miyazono, K, Nagata, K, Saigo, K, Kojima, T, Tanokura, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cooperative DNA-binding and sequence-recognition mechanism of aristaless and clawless
Embo J., 29, 2010
|
|
3MGF
 
 | | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 7.5 | | Descriptor: | Fluorescent protein | | Authors: | Ebisawa, T, Yamamura, A, Ohtsuka, J, Kameda, Y, Hayakawa, K, Nagata, K, Tanokura, M. | | Deposit date: | 2010-04-06 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 7.5
To be Published
|
|
6F2L
 
 | |
4YPJ
 
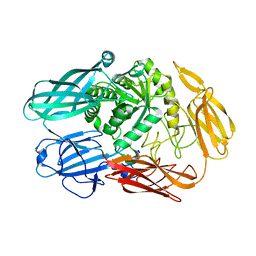 | | X-ray Structure of The Mutant of Glycoside Hydrolase | | Descriptor: | Beta galactosidase | | Authors: | Ishikawa, K, Kataoka, M, Yanamoto, T, Nakabayashi, M, Watanabe, M. | | Deposit date: | 2015-03-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of beta-galactosidase from Bacillus circulans ATCC 31382 (BgaD) and the construction of the thermophilic mutants.
Febs J., 282, 2015
|
|
7BQV
 
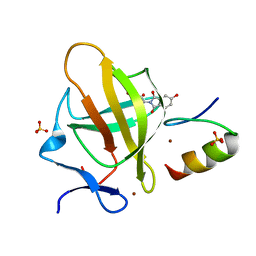 | | Cereblon in complex with SALL4 and (S)-5-hydroxythalidomide | | Descriptor: | 2-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-5-oxidanyl-isoindole-1,3-dione, Protein cereblon, SULFATE ION, ... | | Authors: | Furihata, H, Miyauchi, Y, Asano, A, Tanokura, M, Miyakawa, T. | | Deposit date: | 2020-03-25 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural bases of IMiD selectivity that emerges by 5-hydroxythalidomide.
Nat Commun, 11, 2020
|
|
7BQU
 
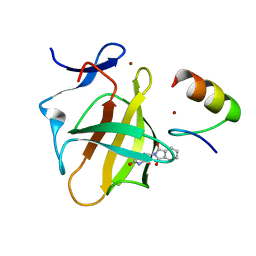 | | Cereblon in complex with SALL4 and (S)-thalidomide | | Descriptor: | Protein cereblon, S-Thalidomide, Sal-like protein 4, ... | | Authors: | Furihata, H, Miyauchi, Y, Asano, A, Tanokura, M, Miyakawa, T. | | Deposit date: | 2020-03-25 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural bases of IMiD selectivity that emerges by 5-hydroxythalidomide.
Nat Commun, 11, 2020
|
|
7ZAV
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA1
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA3
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZAW
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA2
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
3HI1
 
 | | Structure of HIV-1 gp120 (core with V3) in Complex with CD4-Binding-Site Antibody F105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F105 Heavy Chain, F105 Light Chain, ... | | Authors: | Kwon, Y.D, Chen, L, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z, Zhang, M.-Y, Arthos, J, Burton, D.R, Dimitrov, D, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-05-18 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
4XRE
 
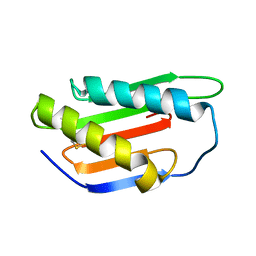 | | Crystal structure of Gnk2 complexed with mannose | | Descriptor: | Antifungal protein ginkbilobin-2, alpha-D-mannopyranose | | Authors: | Miyakawa, T, Hatano, K, Miyauchi, Y, Suwa, Y, Sawano, Y, Tanokura, M. | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity.
Plant Physiol., 166, 2014
|
|
7EKD
 
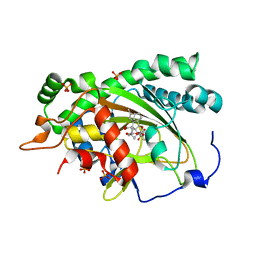 | | Crystal structure of gibberellin 3-oxidase 2 (GA3ox2) in rice | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-OXOGLUTARIC ACID, Gibberellin 3-beta-dioxygenase 2, ... | | Authors: | Takehara, S, Kawai, K, Mikami, B, Ueguchi-Tanaka, M. | | Deposit date: | 2021-04-05 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Evolutionary alterations in gene expression and enzymatic activities of gibberellin 3-oxidase 1 in Oryza.
Commun Biol, 5, 2022
|
|
3IDX
 
 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C222 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab b13 heavy chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
3IDY
 
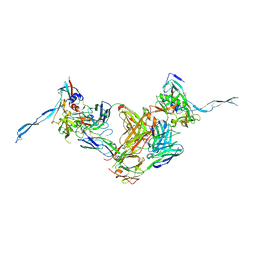 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C2221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab b13 heavy chain, Fab b13 light chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
6KFG
 
 | | Undocked INX-6 hemichannel in detergent | | Descriptor: | Innexin-6 | | Authors: | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2019-07-07 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
3WAZ
 
 | | Crystal structure of a restriction enzyme PabI in complex with DNA | | Descriptor: | ADENINE, DNA (5'-D(*GP*CP*AP*TP*AP*GP*CP*TP*GP*TP*(ORP)P*CP*AP*GP*CP*TP*AP*TP*GP*C)-3'), Putative uncharacterized protein | | Authors: | Miyazono, K, Miyakawa, T, Ito, T, Tanokura, M. | | Deposit date: | 2013-05-10 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A sequence-specific DNA glycosylase mediates restriction-modification in Pyrococcus abyssi.
Nat Commun, 5, 2014
|
|
6KFF
 
 | | Undocked INX-6 hemichannel in a nanodisc | | Descriptor: | Innexin-6 | | Authors: | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2019-07-07 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
3ED1
 
 | | Crystal Structure of Rice GID1 complexed with GA3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GIBBERELLIN A3, Gibberellin receptor GID1, ... | | Authors: | Shimada, A, Nakatsu, T, Ueguchi-Tanaka, M, Kato, H, Matsuoka, M. | | Deposit date: | 2008-09-02 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for gibberellin recognition by its receptor GID1.
Nature, 456, 2008
|
|
3EBL
 
 | | Crystal Structure of Rice GID1 complexed with GA4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GIBBERELLIN A4, Gibberellin receptor GID1, ... | | Authors: | Shimada, A, Nakatsu, T, Ueguchi-Tanaka, M, Kato, H, Matsuoka, M. | | Deposit date: | 2008-08-28 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for gibberellin recognition by its receptor GID1.
Nature, 456, 2008
|
|
3JRS
 
 | | Crystal structure of (+)-ABA-bound PYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At5g46790 | | Authors: | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
3JRQ
 
 | | Crystal structure of (+)-ABA-bound PYL1 in complex with ABI1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Protein phosphatase 2C 56, Putative uncharacterized protein At5g46790 | | Authors: | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
6QVJ
 
 | | HsCKK (human CAMSAP1) decorated 14pf taxol-GDP microtubule | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-03-02 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
6QUY
 
 | | NgCKK (N.Gruberi CKK) decorated 13pf taxol-GDP microtubule | | Descriptor: | CKK domain protein, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
