4YSV
 
 | |
4YSN
 
 | | Structure of aminoacid racemase in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative 4-aminobutyrate aminotransferase | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2015-03-17 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5FB3
 
 | | Structure of glycerophosphate dehydrogenase in complex with NADPH | | Descriptor: | Glycerol-1-phosphate dehydrogenase [NAD(P)+], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYROPHOSPHATE, ... | | Authors: | Sakuraba, H, Hayashi, J, Yamamoto, K, Yoneda, K, Ohshima, T. | | Deposit date: | 2015-12-14 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Unique coenzyme binding mode of hyperthermophilic archaeal sn-glycerol-1-phosphate dehydrogenase from Pyrobaculum calidifontis
Proteins, 84, 2016
|
|
5DM9
 
 | |
5GZ6
 
 | | Structure of D-amino acid dehydrogenase in complex with NADPH and 2-keto-6-aminocapronic acid | | Descriptor: | 6-azanyl-2-oxidanylidene-hexanoic acid, ACETATE ION, Meso-diaminopimelate D-dehydrogenase, ... | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
5GZ3
 
 | | Structure of D-amino acid dehydrogenase in complex with NADP | | Descriptor: | 1,2-ETHANEDIOL, Meso-diaminopimelate D-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
5GZ1
 
 | | Structure of substrate/cofactor-free D-amino acid dehydrogenase | | Descriptor: | Meso-diaminopimelate D-dehydrogenase | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
8HGX
 
 | |
5WYF
 
 | | Structure of amino acid racemase, 2.12 A | | Descriptor: | CADMIUM ION, Isoleucine 2-epimerase, N-[O-PHOSPHONO-PYRIDOXYL]-ISOLEUCINE | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2017-01-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5WYA
 
 | | Structure of amino acid racemase, 2.65 A | | Descriptor: | (2S,3S)-3-methyl-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pentanoic acid, DIMETHYL SULFOXIDE, Isoleucine 2-epimerase | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5H2V
 
 | |
7DV6
 
 | | Discovery of Functionally Selective Transforming Growth Factor beta Type II Receptor (TGF-beta RII) Inhibitors as Anti-Fibrosis Agents | | Descriptor: | 5-[(3S)-5,5-dimethyloxolan-3-yl]-6-methoxy-3-(2-methoxypyridin-4-yl)pyrazolo[1,5-a]pyrimidine, TGF-beta receptor type-2 | | Authors: | Nishihata, J, Nomura, A, Miwa, S, Doi, S, Adachi, T. | | Deposit date: | 2021-01-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of Selective Transforming Growth Factor beta Type II Receptor Inhibitors as Antifibrosis Agents.
Acs Med.Chem.Lett., 12, 2021
|
|
1OUA
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE I56T MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Funahashi, J, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure, stability, and folding process of amyloidogenic mutant human lysozyme.
J.Biochem.(Tokyo), 120, 1996
|
|
2MPH
 
 | |
2LJV
 
 | | Solution structure of Rhodostomin G50L mutant | | Descriptor: | Disintegrin rhodostomin | | Authors: | Chuang, W, Shiu, J, Chen, C, Chen, Y, Chang, Y, Huang, C. | | Deposit date: | 2011-09-29 | | Release date: | 2012-10-03 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Design of Integrin AlphaVbeta3-Specific Disintegrin for Cancer Therapy
To be Published
|
|
3B3M
 
 | | Structure of neuronal NOS heme domain in complex with a inhibitor (+-)-3-{cis-4'-[(6"-aminopyridin-2"-yl)methyl]pyrrolidin-3'-ylamino}propan-1-ol | | Descriptor: | 3-({(3S,4S)-4-[(6-aminopyridin-2-yl)methyl]pyrrolidin-3-yl}amino)propan-1-ol, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Minimal pharmacophoric elements and fragment hopping, an approach directed at molecular diversity and isozyme selectivity. Design of selective neuronal nitric oxide synthase inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
3B3N
 
 | | Structure of neuronal NOS heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-aminopyridin-2"-yl)methyl]pyrrolidin-3'-yl}ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-aminopyridin-2-yl)methyl]pyrrolidin-3-yl}ethane-1,2-diamine, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Minimal pharmacophoric elements and fragment hopping, an approach directed at molecular diversity and isozyme selectivity. Design of selective neuronal nitric oxide synthase inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
2HX4
 
 | | Rat nNOS heme domain complexed with 4-N-(Nw-nitro-L-argininyl)-trans-4-hydroxyamino-L-proline amide | | Descriptor: | (4R)-4-(HYDROXY{N~5~-[IMINO(NITROAMINO)METHYL]-L-ORNITHYL}AMINO)-L-PROLINAMIDE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2006-08-02 | | Release date: | 2007-04-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design and Synthesis of N(omega)-Nitro-l-Arginine-Containing Peptidomimetics as Selective Inhibitors of Neuronal Nitric Oxide Synthase. Displacement of the Heme Structural Water.
J.Med.Chem., 50, 2007
|
|
2HX2
 
 | | Bovine eNOS heme domain complexed with (4S)-N-{4-Amino-5-[(2-aminoethyl)-hydroxyamino]-pentyl}-N'-nitroguanidine | | Descriptor: | (4S)-N-{4-AMINO-5-[(2-AMINOETHYL)(HYDROXYAMINO]-PENTYL}-N'-NITROGUANIDINE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2006-08-02 | | Release date: | 2007-04-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design and Synthesis of N(omega)-Nitro-l-Arginine-Containing Peptidomimetics as Selective Inhibitors of Neuronal Nitric Oxide Synthase. Displacement of the Heme Structural Water.
J.Med.Chem., 50, 2007
|
|
2HX3
 
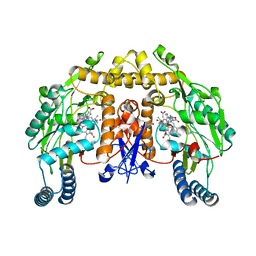 | | Rat nNOS heme domain complexed with (4S)-N-{4-Amino-5-[(2-aminoethyl)-hydroxyamino]-pentyl}-N'-nitroguanidine | | Descriptor: | (4S)-N-{4-AMINO-5-[(2-AMINOETHYL)(HYDROXYAMINO]-PENTYL}-N'-NITROGUANIDINE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2006-08-02 | | Release date: | 2007-04-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design and Synthesis of N(omega)-Nitro-l-Arginine-Containing Peptidomimetics as Selective Inhibitors of Neuronal Nitric Oxide Synthase. Displacement of the Heme Structural Water.
J.Med.Chem., 50, 2007
|
|
5K79
 
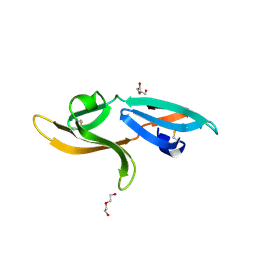 | | Structure and anti-HIV activity of CYT-CVNH, a new cyanovirin-n homolog | | Descriptor: | 1,2-ETHANEDIOL, Cyanovirin-N domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Matei, E, Basu, R, Furey, W, Shi, J, Calnan, C, Aiken, C, Gronenborn, A.M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Glycan Binding of a New Cyanovirin-N Homolog.
J.Biol.Chem., 291, 2016
|
|
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
6LNU
 
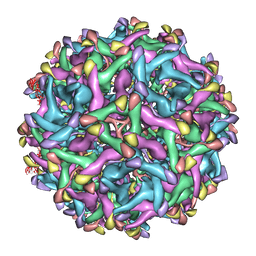 | | Cryo-EM structure of immature Zika virus | | Descriptor: | Genome polyprotein | | Authors: | Tan, T.Y, Fibriansah, G, Kostyuchenko, V.A, Ng, T.S, Lim, X.X, Lim, X.N, Shi, J, Morais, M.C, Corti, D, Lok, S.M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Capsid protein structure in Zika virus reveals the flavivirus assembly process.
Nat Commun, 11, 2020
|
|
5X8R
 
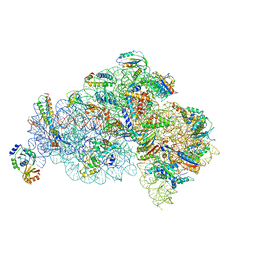 | | Structure of the 30S small subunit of chloroplast ribosome from spinach | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, chloroplastic, ... | | Authors: | Ahmed, T, Shi, J, Bhushan, S. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation
Nucleic Acids Res., 45, 2017
|
|
5X8P
 
 | | Structure of the 70S chloroplast ribosome from spinach | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Ahmed, T, Shi, J, Bhushan, S. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation
Nucleic Acids Res., 45, 2017
|
|
