7TF3
 
 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) Q484A spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
7TF5
 
 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
7TF1
 
 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) Q484I spike protein (focused refinement of RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
7TF4
 
 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) spike protein (focused refinement of RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
7KIY
 
 | | Plasmodium falciparum RhopH complex in soluble form | | Descriptor: | Cytoadherence linked asexual protein 3, High molecular weight rhoptry protein 3, High molecular weight rhoptry protein-2 | | Authors: | Schureck, M.A, Darling, J.E, Merk, A, Subramaniam, S, Desai, S.A. | | Deposit date: | 2020-10-25 | | Release date: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Malaria parasites use a soluble RhopH complex for erythrocyte invasion and an integral form for nutrient uptake.
Elife, 10, 2021
|
|
6B44
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound target dsDNA | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy2, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B45
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy2, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B47
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF2 | | Descriptor: | Anti-CRISPR protein AcrF2, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B48
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF10 | | Descriptor: | Anti-CRISPR protein AcrF10, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B46
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF1 | | Descriptor: | Anti-CRISPR protein AcrF1, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy3, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
8DI5
 
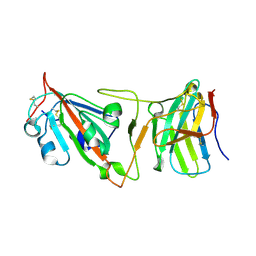 | | Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein in complex with VH domain F6 (focused refinement of RBD and VH F6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH F6 | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Subramaniam, S. | | Deposit date: | 2022-06-28 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Potent and broad neutralization of SARS-CoV-2 variants of concern (VOCs) including omicron sub-lineages BA.1 and BA.2 by biparatopic human VH domains.
Iscience, 25, 2022
|
|
7SZ7
 
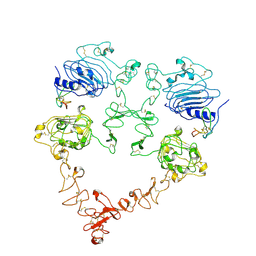 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to TGF-alpha. "tips-juxtaposed" conformation | | Descriptor: | Epidermal growth factor receptor, Transforming growth factor alpha | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SZ1
 
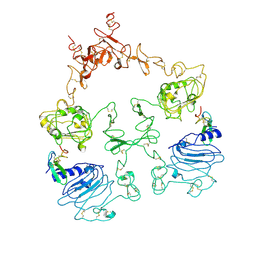 | | Cryo-EM structure of the extracellular module of the full-length EGFR L834R bound to EGF. "tips-separated" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SYD
 
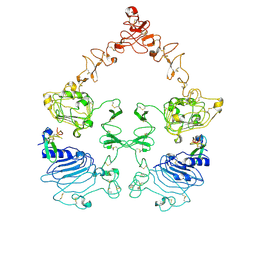 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to EGF "tips-juxtaposed" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SZ5
 
 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to TGF-alpha "tips-separated" conformation | | Descriptor: | Epidermal growth factor receptor, Transforming growth factor alpha | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SYE
 
 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to EGF. "tips-separated" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SZ0
 
 | | Cryo-EM structure of the extracellular module of the full-length EGFR L834R bound to EGF. "tips-juxtaposed" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7T9J
 
 | | Cryo-EM structure of the SARS-CoV-2 Omicron spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2021-12-19 | | Release date: | 2021-12-29 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Science, 375, 2022
|
|
7T9K
 
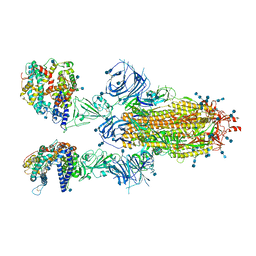 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2021-12-19 | | Release date: | 2021-12-29 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Science, 375, 2022
|
|
7T9L
 
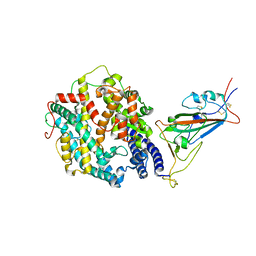 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2021-12-19 | | Release date: | 2021-12-29 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Science, 375, 2022
|
|
4ABN
 
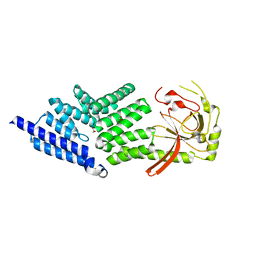 | | Crystal structure of full length mouse Strap (TTC5) | | Descriptor: | 1,2-ETHANEDIOL, TETRATRICOPEPTIDE REPEAT PROTEIN 5 | | Authors: | Pike, A.C.W, Bullock, A.N, Kleinekofort, W, Zimmermann, T, Burgess-Brown, N, Sharpe, T.D, Thangaratnarajah, C, Keates, T, Ugochukwu, E, Bunkoczi, G, Uppenberg, J, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, La Thangue, N.B, Knapp, S. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The P53 Cofactor Strap Exhibits an Unexpected Tpr Motif and Oligonucleotide-Binding (Ob)-Fold Structure.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7MEQ
 
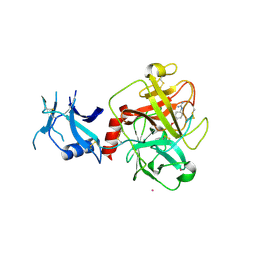 | | Crystal structure of human TMPRSS2 in complex with Nafamostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, Transmembrane protease serine 2, ... | | Authors: | Fraser, B, Beldar, S, Hutchinson, A, Li, Y, Seitova, A, Edwards, A.M, Benard, F, Arrowsmith, C.H, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-07 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation.
Nat.Chem.Biol., 18, 2022
|
|
7MBO
 
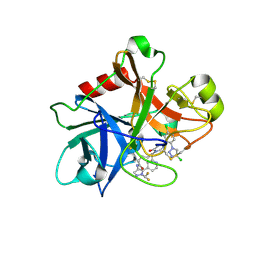 | | FACTOR XIA (PICHIA PASTORIS; C500S [C122S]) IN COMPLEX WITH THE INHIBITOR Milvexian (BMS-986177), IUPAC NAME:(6R,10S)-10-{4-[5-chloro-2-(4-chloro-1H-1,2,3-triazol-1-yl)phenyl]-6- oxopyrimidin-1(6H)-yl}-1-(difluoromethyl)-6-methyl-1,4,7,8,9,10-hexahydro-15,11- (metheno)pyrazolo[4,3-b][1,7]diazacyclotetradecin-5(6H)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa light chain, Milvexian | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.924 Å) | | Cite: | Discovery of Milvexian, a High-Affinity, Orally Bioavailable Inhibitor of Factor XIa in Clinical Studies for Antithrombotic Therapy.
J.Med.Chem., 65, 2022
|
|
6W50
 
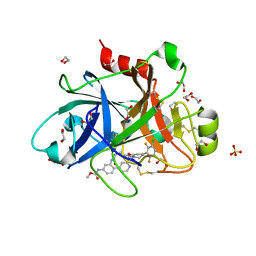 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR METHYL ((10R,14S)- 14-(4-(3-CHLORO-2,6-DIFLUOROPHENYL)-6-OXO-3,6-DIHYDRO- 1(2H)-PYRIDINYL)-10-METHYL-9-OXO-8,16- DIAZATRICYCLO[13.3.1.0~2,7~]NONADECA-1(19),2,4,6,15,17- HEXAEN-5-YL)CARBAMATE | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2020-03-12 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a High Affinity, Orally Bioavailable Macrocyclic FXIa Inhibitor with Antithrombotic Activity in Preclinical Species.
J.Med.Chem., 63, 2020
|
|
4ZDO
 
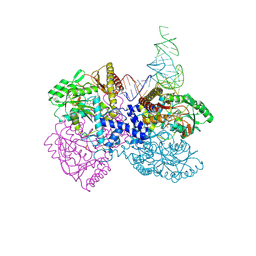 | |
