7JO1
 
 | | Carbonic Anhydrase IX Mimic Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)butyramide | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)butanamide, ... | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
7JOB
 
 | | Carbonic Anhydrase IX Mimic Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)cyclohexanecarboxamide | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)cyclohexanecarboxamide, ... | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-06 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
7K6K
 
 | | Carbonic Anhydrase II complexed with 4-(2-(3-(3,5-dimethylphenyl)ureido)ethylsulfonamido)benzenesulfonamide | | Descriptor: | 4-{[(2-{[(3,5-dimethylphenyl)carbamoyl]amino}ethyl)sulfonyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Andring, J.T, Singh, S, McKenna, R. | | Deposit date: | 2020-09-20 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.306 Å) | | Cite: | Handling drug-target selectivity: A study on ureido containing Carbonic Anhydrase inhibitors.
Eur.J.Med.Chem., 212, 2021
|
|
7K6T
 
 | | Carbonic Anhydrase IX mimic complexed with 4-(2-(3-phenylureido)ethylsulfonamido)benzenesulfonamide | | Descriptor: | 4-[({2-[(phenylcarbamoyl)amino]ethyl}sulfonyl)amino]benzene-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Andring, J.T, Singh, S, McKenna, R. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Handling drug-target selectivity: A study on ureido containing Carbonic Anhydrase inhibitors.
Eur.J.Med.Chem., 212, 2021
|
|
7K6J
 
 | | Carbonic Anhydrase II complexed with 4-(2-(3-(4-methylphenyl)ureido)ethylsulfonamido)benzenesulfonamide | | Descriptor: | 4-{[(2-{[(4-methylphenyl)carbamoyl]amino}ethyl)sulfonyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Andring, J.T, Singh, S, McKenna, R. | | Deposit date: | 2020-09-20 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Handling drug-target selectivity: A study on ureido containing Carbonic Anhydrase inhibitors.
Eur.J.Med.Chem., 212, 2021
|
|
7K6U
 
 | | Carbonic Anhydrase IX mimic complexed with 4-(2-(3-(4-methylphenyl)ureido)ethylsulfonamido)benzenesulfonamide | | Descriptor: | 4-{[(2-{[(4-methylphenyl)carbamoyl]amino}ethyl)sulfonyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Andring, J.T, Singh, S, McKenna, R. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Handling drug-target selectivity: A study on ureido containing Carbonic Anhydrase inhibitors.
Eur.J.Med.Chem., 212, 2021
|
|
7K6L
 
 | |
7JOC
 
 | | Carbonic Anhydrase IX Mimic Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)adamantanecarboxamide | | Descriptor: | (3S,5S,7S)-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)tricyclo[3.3.1.1~3,7~]decane-1-carboxamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-06 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.385 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
7K6Z
 
 | | Carbonic Anhydrase IX mimic complexed with 4-(2-(3-(4-fluorophenyl)ureido)ethylsulfonamido)benzenesulfonamide | | Descriptor: | 4-{[(2-{[(4-fluorophenyl)carbamoyl]amino}ethyl)sulfonyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Andring, J.T, Singh, S, McKenna, R. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | Handling drug-target selectivity: A study on ureido containing Carbonic Anhydrase inhibitors.
Eur.J.Med.Chem., 212, 2021
|
|
2LNL
 
 | | Structure of human CXCR1 in phospholipid bilayers | | Descriptor: | C-X-C chemokine receptor type 1 | | Authors: | Park, S, Das, B.B, Casagrande, F, Nothnagel, H, Chu, M, Kiefer, H, Maier, K, De Angelis, A, Marassi, F.M, Opella, S.J. | | Deposit date: | 2011-12-31 | | Release date: | 2012-10-17 | | Last modified: | 2016-04-27 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the chemokine receptor CXCR1 in phospholipid bilayers.
Nature, 491, 2012
|
|
4IE9
 
 | | Bovine PKA C-alpha in complex with 3-pyridylmethyl-5-methyl-1H-pyrazole-3-carboxylate | | Descriptor: | GLYCEROL, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha, ... | | Authors: | Dreyer, M.K, Schiffer, A, Lodge, J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-05-01 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Accounting for Conformational Variability in Protein-Ligand Docking with NMR-Guided Rescoring
J.Am.Chem.Soc., 135, 2013
|
|
4IJ9
 
 | | Bovine PKA C-alpha in complex with 2-[[5-(4-pyridyl)-1H-1,2,4-triazol-3-yl]sulfanyl]-1-(2-thiophenyl)ethanone | | Descriptor: | 2-[[5-(4-pyridyl)-1H-1,2,4-triazol-3-yl]sulfanyl]-1-(2-thiophenyl)ethanone, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Dreyer, M.K, Schiffer, A. | | Deposit date: | 2012-12-21 | | Release date: | 2013-05-01 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Accounting for Conformational Variability in Protein-Ligand Docking with NMR-Guided Rescoring
J.Am.Chem.Soc., 135, 2013
|
|
4XT3
 
 | | Structure of a viral GPCR bound to human chemokine CX3CL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fractalkine, G-protein coupled receptor homolog US28, ... | | Authors: | Burg, J.S, Jude, K.M, Waghray, D, Garcia, K.C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor.
Science, 347, 2015
|
|
4XT1
 
 | | Structure of a nanobody-bound viral GPCR bound to human chemokine CX3CL1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Fractalkine, ... | | Authors: | Burg, J.S, Jude, K.M, Waghray, D, Garcia, K.C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor.
Science, 347, 2015
|
|
5IFH
 
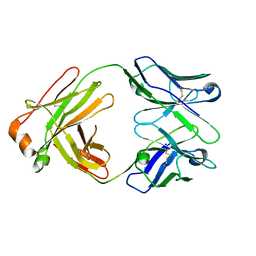 | | Crystal structure of the BCR Fab fragment from subset #2 case P11475 | | Descriptor: | Heavy chain of the Fab fragment from BCR derived from the P11475 CLL clone, Light chain of the Fab fragment from BCR derived from the P11475 CLL clone | | Authors: | Minici, C, Degano, M. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Distinct homotypic B-cell receptor interactions shape the outcome of chronic lymphocytic leukaemia.
Nat Commun, 8, 2017
|
|
5DRX
 
 | |
6ODL
 
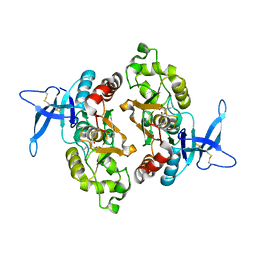 | | Crystal structure of GluN2A agonist binding domain with 4-butyl-(S)-CCG-IV | | Descriptor: | (1S,2R)-2-[(S)-amino(carboxy)methyl]-1-butylcyclopropane-1-carboxylic acid, Glutamate receptor ionotropic, NMDA 2A,Glutamate receptor ionotropic, ... | | Authors: | Mou, T.C, Clausen, R.P, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stereoselective synthesis of novel 2'-(S)-CCG-IV analogues as potent NMDA receptor agonists.
Eur.J.Med.Chem., 212, 2021
|
|
5DRW
 
 | |
5J74
 
 | | Fluorogen activating protein AM2.2 in complex with TO1-2p | | Descriptor: | 1-(17-amino-5,8-dioxo-12,15-dioxa-4,9-diazaheptadecan-1-yl)-4-{[3-(3-sulfopropyl)-1,3-benzothiazol-3-ium-2-yl]methyl}quinolin-1-ium, PHOSPHATE ION, scFv AM2.2 | | Authors: | Stanfield, R.L, Wilson, I.A, Wu, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Small-Molecule Nonfluorescent Inhibitors of Fluorogen-Fluorogen Activating Protein Binding Pair.
J Biomol Screen, 21, 2016
|
|
5J75
 
 | | Fluorogen Activating Protein AM2.2 in complex with ML342 | | Descriptor: | N,4-dimethyl-N-{2-oxo-2-[4-(pyridin-2-yl)piperazin-1-yl]ethyl}benzene-1-sulfonamide, PHOSPHATE ION, scFv AM2.2 | | Authors: | Stanfield, R.L, Wilson, I.A, Wu, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Small-Molecule Nonfluorescent Inhibitors of Fluorogen-Fluorogen Activating Protein Binding Pair.
J Biomol Screen, 21, 2016
|
|
6ZTD
 
 | | Crystal structure of the BCR Fab fragment from subset #169 case P6540 | | Descriptor: | Heavy chain of the Fab fragment from BCR derived from the P6540 CLL clone, Light chain of the Fab fragment from BCR derived from the P6540 CLL clone | | Authors: | Carriles, A.A, Minici, C, Degano, M. | | Deposit date: | 2020-07-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Higher-order immunoglobulin repertoire restrictions in CLL: the illustrative case of stereotyped subsets 2 and 169.
Blood, 137, 2021
|
|
1NE5
 
 | | Solution Structure of HERG Specific Scorpion Toxin CnErg1 | | Descriptor: | ergtoxin | | Authors: | Torres, A.M, Paramjit, B, Alewood, P, Kuchel, P.W, Vandenberg, J.I. | | Deposit date: | 2002-12-10 | | Release date: | 2003-04-01 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of CnErg1 (Ergtoxin), a HERG specific scorpion toxin
FEBS Lett., 539, 2003
|
|
4MNV
 
 | | Crystal structure of bicyclic peptide UK729 bound as an acyl-enzyme intermediate to urokinase-type plasminogen activator (uPA) | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, ACETATE ION, GLYCEROL, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide ligands stabilized by small molecules.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4MNX
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK811 | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Peptide ligands stabilized by small molecules.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4MNY
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK903 | | Descriptor: | ACETATE ION, GLYCEROL, N,N',N''-benzene-1,3,5-triyltris(2-bromoacetamide), ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Peptide ligands stabilized by small molecules.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
