4NV3
 
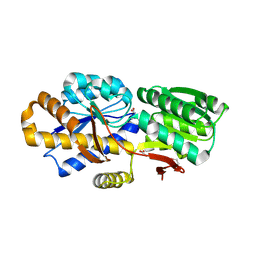 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with valine. | | Descriptor: | ACETATE ION, Amino acid/amide ABC transporter substrate-binding protein, HAAT family, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-12-04 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with valine.
To be Published
|
|
2OGG
 
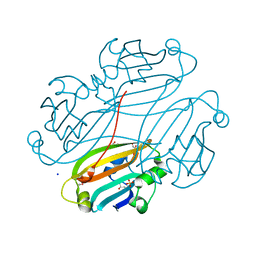 | | Structure of B. subtilis trehalose repressor (TreR) effector binding domain | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Rezacova, P, Krejcirikova, V, Borek, D, Moy, S.F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the effector-binding domain of the trehalose repressor TreR from Bacillus subtilis 168 reveals a unique quarternary assembly.
Proteins, 69, 2007
|
|
2OKU
 
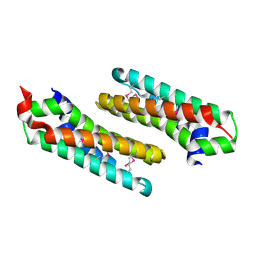 | |
4Q29
 
 | | Ensemble Refinement of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 protein | | Authors: | Wang, F, Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Miller, M.D, Thomas, M.G, Joachimiak, A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-07 | | Last modified: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
2O3G
 
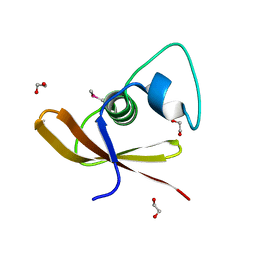 | | Structural Genomics, the crystal structure of a conserved putative domain from Neisseria meningitidis MC58 | | Descriptor: | 1,2-ETHANEDIOL, Putative protein | | Authors: | Tan, K, Volkart, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of a conserved putative domain from Neisseria meningitidis MC58
To be Published
|
|
4Q88
 
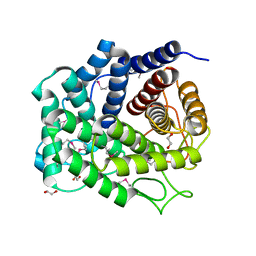 | | Glycosyl hydrolase family 88 from Bacteroides vulgatus | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uncharacterized protein | | Authors: | Osipiuk, J, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Glycosyl hydrolase Family 88 from Bacteroides vulgatus
To be Published
|
|
4QM1
 
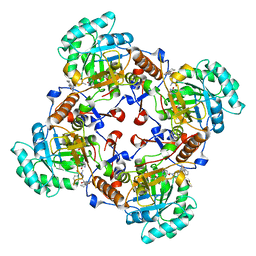 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor D67 | | Descriptor: | 2-(3-methyl-4-oxo-3,4-dihydrophthalazin-1-yl)-N-(6,7,8,9-tetrahydrodibenzo[b,d]furan-2-yl)acetamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Mandapati, K, Gollapalli, D, Gorla, S.K, Zhang, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-14 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7964 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor D67
To be Published, 2014
|
|
4PZK
 
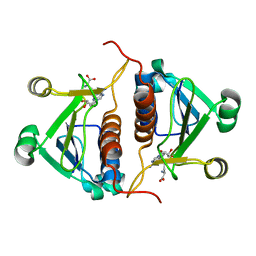 | | Crystal strucrure of putative RNA methyltransferase from Bacillus anthracis. | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (cytidine(34)-2'-O)-methyltransferase | | Authors: | Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-31 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal strucrure of putative RNA methyltransferase from Bacillus anthracis.
To be Published
|
|
3D1R
 
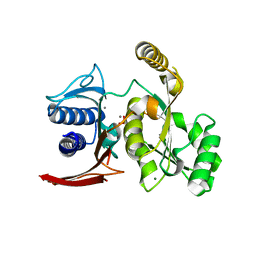 | | Structure of E. coli GlpX with its substrate fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Singer, A, Skarina, T, Dong, A, Brown, G, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
2OCD
 
 | | Crystal structure of L-asparaginase I from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | ACETATE ION, GLYCEROL, L-asparaginase I | | Authors: | Nocek, B, Wu, R, Osipiuk, J, Moy, S, Kim, Y, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-20 | | Release date: | 2007-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of L-asparaginase I from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
4O5A
 
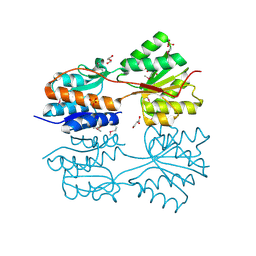 | | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140 | | Descriptor: | GLYCEROL, LacI family transcription regulator, SULFATE ION | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140.
To be Published
|
|
2OBB
 
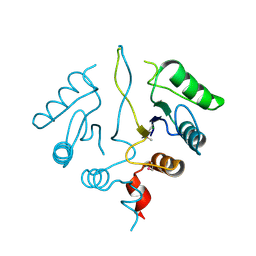 | |
2OEE
 
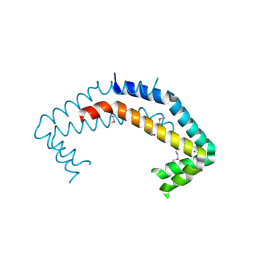 | |
2NR7
 
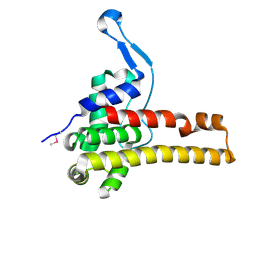 | | Structural Genomics, the crystal structure of putative secretion activator protein from Porphyromonas gingivalis W83 | | Descriptor: | Secretion activator protein, putative | | Authors: | Tan, K, Bigelow, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-01 | | Release date: | 2006-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The crystal structure of putative secretion activator protein from Porphyromonas gingivalis W83
To be Published
|
|
2NRK
 
 | |
4PIB
 
 | | Crystal Structure of Uncharacterized Conserved Protein PixA from Burkholderia thailandensis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-05-08 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Uncharacterized Conserved Protein PixA from Burkholderia thailandensis
To Be Published
|
|
4PDY
 
 | | Crystal structure of aminoglycoside phosphotransferase from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | Aminoglycoside phosphotransferase, HISTIDINE, THIOCYANATE ION | | Authors: | Chang, C, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-22 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of aminoglycoside phosphotransferase from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446
To be published
|
|
2OD0
 
 | | The crystal structure of gene product VP1028 from Vibrio parahaemolyticus | | Descriptor: | Hypothetical protein VP1028, MAGNESIUM ION, SULFATE ION | | Authors: | Zhang, R, Zhou, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-21 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of gene product VP1028 from Vibrio parahaemolyticus
To be Published
|
|
2OCH
 
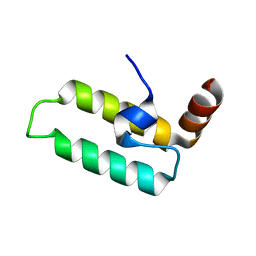 | | J-domain of dnj-12 from Caenorhabditis elegans | | Descriptor: | Hypothetical protein dnj-12 | | Authors: | Osipiuk, J, Maltseva, N, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-20 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | X-ray crystal structure of J-domain of dnj-12 from Caenorhabditis elegans.
To be Published
|
|
4QJG
 
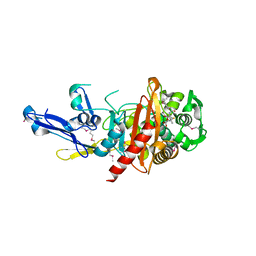 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with penicillin V | | Descriptor: | (2R,4S)-5,5-dimethyl-2-{(1R)-2-oxo-1-[(phenoxyacetyl)amino]ethyl}-1,3-thiazolidine-4-carboxylic acid, Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-06-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with penicillin V
To be Published
|
|
4PYR
 
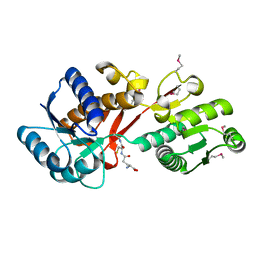 | | Structure of a putative branched-chain amino acid ABC transporter from Chromobacterium violaceum ATCC 12472 | | Descriptor: | GLUTATHIONE, Putative branched-chain amino acid ABC transporter | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-27 | | Release date: | 2014-04-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a putative branched-chain amino acid ABC transporter from Chromobacterium violaceum ATCC 12472
To be Published
|
|
2O3F
 
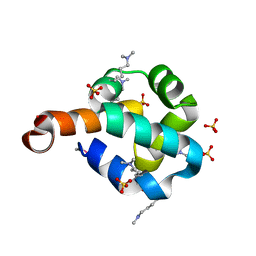 | | Structural Genomics, the crystal structure of the N-terminal domain of the putative transcriptional regulator ybbH from Bacillus subtilis subsp. subtilis str. 168. | | Descriptor: | Putative HTH-type transcriptional regulator ybbH, SULFATE ION | | Authors: | Tan, K, Bigelow, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the N-terminal domain of the putative transcriptional regulator ybbH from Bacillus subtilis subsp. subtilis str. 168.
To be Published
|
|
4Q31
 
 | | The crystal structure of cystathione gamma lyase (CalE6) from Micromonospora echinospora | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-07 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural dynamics of a methionine gamma-lyase for calicheamicin biosynthesis: Rotation of the conserved tyrosine stacking with pyridoxal phosphate.
Struct Dyn, 3, 2016
|
|
4QA9
 
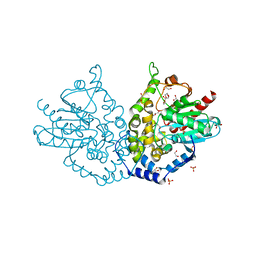 | | Ensemble refinement of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | 1,2-ETHANEDIOL, Epoxide hydrolase, SULFATE ION | | Authors: | Wang, F, Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Joachimiak, A, Phillips Jr, G.N, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Ensemble refinement of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
4Q6B
 
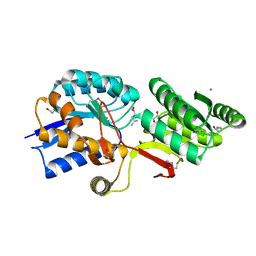 | | Crystal Structure of ABC Transporter Substrate-Binding Protein fromDesulfitobacterium hafniense complex with Leu | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Extracellular ligand-binding receptor, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-22 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.667 Å) | | Cite: | Crystal Structure of ABC Transporter Substrate-Binding Protein fromDesulfitobacterium hafniense complex with Leu
To be Published, 2014
|
|
