4LWO
 
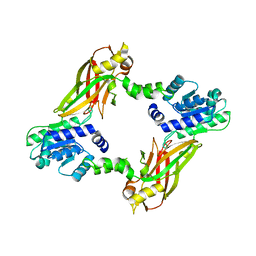 | | Crystal structure of PRMT6 | | Descriptor: | Arginine N-methyltransferase, putative | | Authors: | Zhu, Y, Wang, C, Shi, Y, Teng, M. | | Deposit date: | 2013-07-28 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal Structure of Arginine Methyltransferase 6 from Trypanosoma brucei
Plos One, 9, 2014
|
|
4M51
 
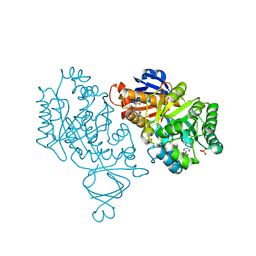 | | Crystal structure of amidohydrolase nis_0429 (ser145ala mutant) from nitratiruptor sp. sb155-2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Amidohydrolase family protein, BENZOIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Gobble, A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-07 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Deamination of 6-aminodeoxyfutalosine in menaquinone biosynthesis by distantly related enzymes.
Biochemistry, 52, 2013
|
|
4M13
 
 | |
4LWS
 
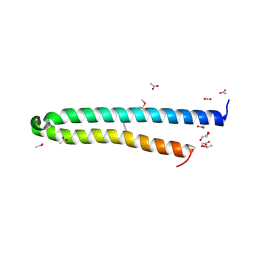 | | EsxA : EsxB (SeMet) hetero-dimer from Thermomonospora curvata | | Descriptor: | ACETATE ION, GLYCEROL, Uncharacterized protein | | Authors: | Dovala, D.L, Cox, J.S, Stroud, R.M, Rosenberg, O.S. | | Deposit date: | 2013-07-28 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrates Control Multimerization and Activation of the Multi-Domain ATPase Motor of Type VII Secretion.
Cell(Cambridge,Mass.), 161, 2015
|
|
4M0Z
 
 | |
4M12
 
 | |
4MBJ
 
 | |
4MKI
 
 | | Cobalt transporter ATP-binding subunit | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Energy-coupling factor transporter ATP-binding protein EcfA2, SULFATE ION | | Authors: | Yu, Y, Zhang, L, Chai, C.L, Heymann, D. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for a homodimeric ATPase subunit of an ECF transporter
Protein Cell, 4, 2013
|
|
5H5Z
 
 | | Crystal structure of bony fish MHC class I, peptide and B2m II | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide chain | | Authors: | Chen, Z, Zhang, N, Qi, J, Li, X, Chen, R, Wang, Z, Gao, F.G, Xia, C. | | Deposit date: | 2016-11-10 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Mechanism of beta 2m Molecule-Induced Changes in the Peptide Presentation Profile in a Bony Fish.
Iscience, 23, 2020
|
|
4MWH
 
 | | Crystal structure of scCK2 alpha in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, MAGNESIUM ION, ... | | Authors: | Liu, H. | | Deposit date: | 2013-09-24 | | Release date: | 2013-11-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NUX
 
 | | Structure of receptor A | | Descriptor: | Interleukin-17 receptor A | | Authors: | Zhang, B, Han, Y, Deng, J. | | Deposit date: | 2013-12-04 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Structure of the unique SEFIR domain from human interleukin 17 receptor A reveals a composite ligand-binding site containing a conserved alpha-helix for Act1 binding and IL-17 signaling.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7VOJ
 
 | | Al-bound structure of the AtALMT1 mutant M60A | | Descriptor: | ACETIC ACID, ALUMINUM ION, Aluminum-activated malate transporter 1 | | Authors: | Wang, J. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ALMT1-mediated aluminum resistance in Arabidopsis.
Cell Res., 32, 2022
|
|
7VQ3
 
 | |
7VQ4
 
 | |
7VQ7
 
 | | The Al-bound AtALMT1 structure at pH 5 (ALMT1Al/pH5) | | Descriptor: | ALUMINUM ION, Aluminum-activated malate transporter 1 | | Authors: | Wang, J.Q. | | Deposit date: | 2021-10-19 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of ALMT1-mediated aluminum resistance in Arabidopsis.
Cell Res., 32, 2022
|
|
7VQ5
 
 | |
7VCF
 
 | | Cryo-EM structure of Chlamydomonas TOC-TIC supercomplex | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wu, J, Yan, Z, Jin, Z, Zhang, Y. | | Deposit date: | 2021-09-02 | | Release date: | 2022-11-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of a TOC-TIC supercomplex spanning two chloroplast envelope membranes.
Cell, 185, 2022
|
|
4LYA
 
 | | EssC (ATPases 2 and 3) from Geobacillus thermodenitrificans (SeMet) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Uncharacterized protein | | Authors: | Dovala, D.L, Bendebury, A, Cox, J.S, Stroud, R.M, Rosenberg, O.S. | | Deposit date: | 2013-07-30 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Substrates Control Multimerization and Activation of the Multi-Domain ATPase Motor of Type VII Secretion.
Cell(Cambridge,Mass.), 161, 2015
|
|
4M14
 
 | |
4M15
 
 | |
6K2Z
 
 | | Human Galectin-14 with lactose | | Descriptor: | Placental protein 13-like, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2019-05-15 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-function studies of galectin-14, an important effector molecule in embryology.
Febs J., 288, 2021
|
|
7XK2
 
 | | Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, DiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
4OK6
 
 | |
4OK3
 
 | |
8Z8Z
 
 | | Cryo-EM structure of LYCHOS | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Xiong, Q, Zhu, Z, Li, T, Zhou, Z, Chao, Y, Qu, Q, Li, D. | | Deposit date: | 2024-04-22 | | Release date: | 2025-01-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Molecular architecture of human LYCHOS involved in lysosomal cholesterol signaling.
Nat.Struct.Mol.Biol., 32, 2025
|
|
