7X37
 
 | | Cryo-EM structure of Coxsackievirus B1 A particle in complex with nAb 2E6 (CVB1-A:2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3E
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A-particle in complex with nAb 9A3 (CVB1-pre-A:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2G
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb nAb 2E6 (CVB1-E:2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-25 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2O
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 2E6 (CVB1-M:2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-25 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X49
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 9A3) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3D
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 9A3 (CVB1-M:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
4OU4
 
 | | Crystal structure of esterase rPPE mutant S159A complexed with (S)-Ac-CPA | | Descriptor: | (2S)-(acetyloxy)(2-chlorophenyl)ethanoic acid, Alpha/beta hydrolase fold-3 domain protein | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-02-15 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4OB7
 
 | | Crystal structure of esterase rPPE mutant W187H | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4ORD
 
 | |
4OU5
 
 | | Crystal structure of esterase rPPE mutant S159A/W187H | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-02-15 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
7X40
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X46
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 2E6 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X47
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 2E6 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, Genome polyprotein, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3C
 
 | | Cryo-EM structure of Coxsackievirus B1 muture virion in complex with nAbs 8A10 and 5F5 (CVB1-M:8A10:5F5) | | Descriptor: | 5F5 heavy chain, 5F5 light chain, 8A10 heavy chain, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X4K
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (classified from CVB1 mature virion in complex with 8A10 and 9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, Genome polyprotein, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X4M
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10, 2E6 and 9A3) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
4OB6
 
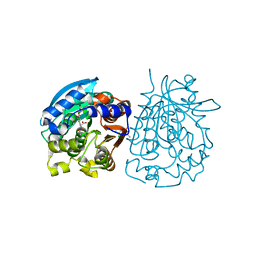 | | Complex structure of esterase rPPE S159A/W187H and substrate (S)-Ac-CPA | | Descriptor: | (2S)-(acetyloxy)(2-chlorophenyl)ethanoic acid, Alpha/beta hydrolase fold-3 domain protein | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Chen, Q, Zhou, J.H, Xu, J.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
7VXX
 
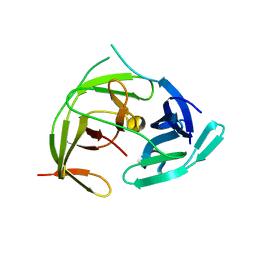 | | Zika virus NS2B/NS3 protease bZipro(C143S) in complex with 4-amino benzamidine | | Descriptor: | P-AMINO BENZAMIDINE, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Xiong, Y.C, Cheng, F, Zhang, J.Y, Su, H.X, Hu, H.C, Zou, Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2021-11-13 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design of a novel inhibitor of the ZIKA virus NS2B/NS3 protease.
Bioorg.Chem., 128, 2022
|
|
7VXY
 
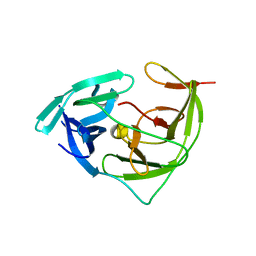 | | Zika virus NS2B/NS3 protease bZipro(C143S) in complex with D-RKOR | | Descriptor: | Peptide inhibitor, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Xiong, Y.C, Cheng, F, Zhang, J.Y, Su, H.X, Hu, H.C, Zou, Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2021-11-13 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure-based design of a novel inhibitor of the ZIKA virus NS2B/NS3 protease.
Bioorg.Chem., 128, 2022
|
|
5Y18
 
 | |
3ROC
 
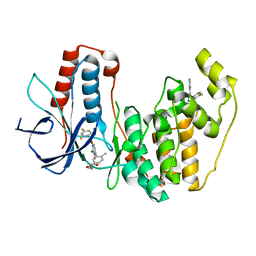 | | Crystal structure of human p38 alpha complexed with a pyrimidinone compound | | Descriptor: | 3-{5-chloro-4-[(2,4-difluorobenzyl)oxy]-6-oxopyrimidin-1(6H)-yl}-N-(2-hydroxyethyl)-4-methylbenzamide, 4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Shieh, H.-S, Xing, L. | | Deposit date: | 2011-04-25 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substituted N-aryl-6-pyrimidinones: A new class of potent, selective, and orally active p38 MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7XY7
 
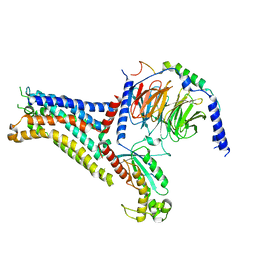 | | Adenosine receptor bound to a non-selective agonist in complex with a G protein obtained by cryo-EM | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Zhang, J.Y, Chen, Y, Hua, T, Song, G.J. | | Deposit date: | 2022-05-31 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Cryo-EM structure of the human adenosine A 2B receptor-G s signaling complex.
Sci Adv, 8, 2022
|
|
7XY6
 
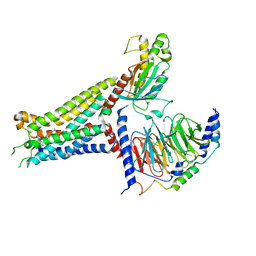 | | Adenosine receptor bound to an agonist in complex with G protein obtained by cryo-EM | | Descriptor: | 2-[6-azanyl-3,5-dicyano-4-[4-(cyclopropylmethoxy)phenyl]pyridin-2-yl]sulfanylethanamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, J.Y, Chen, Y, Hua, T, Song, G.J. | | Deposit date: | 2022-05-31 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of the human adenosine A 2B receptor-G s signaling complex.
Sci Adv, 8, 2022
|
|
5ZLF
 
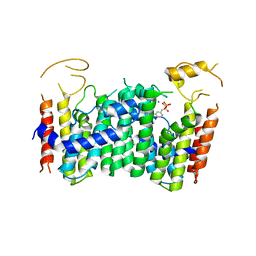 | | CRYSTAL STRUCTURE OF OCTAPRENYL PYROPHOSPHATE SYNTHASE FROM ESCHERICHIA COLI WITH ligand BPH-629 | | Descriptor: | MAGNESIUM ION, Octaprenyl diphosphate synthase, [2-(3-DIBENZOFURAN-4-YL-PHENYL)-1-HYDROXY-1-PHOSPHONO-ETHYL]-PHOSPHONIC ACID | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-03-27 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | Discovery of Lipophilic Bisphosphonates That Target Bacterial Cell Wall and Quinone Biosynthesis.
J.Med.Chem., 62, 2019
|
|
5ZE6
 
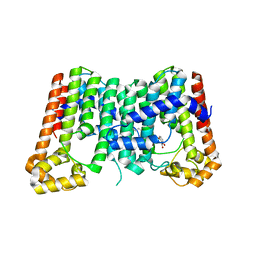 | | CRYSTAL STRUCTURE OF OCTAPRENYL PYROPHOSPHATE SYNTHASE FROM ESCHERICHIA COLI WITH BPH-981 | | Descriptor: | 2-hydroxy-6-(tetradecyloxy)benzoic acid, MAGNESIUM ION, Octaprenyl diphosphate synthase | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-02-26 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Lipophilic Bisphosphonates That Target Bacterial Cell Wall and Quinone Biosynthesis.
J.Med.Chem., 62, 2019
|
|
