4MY8
 
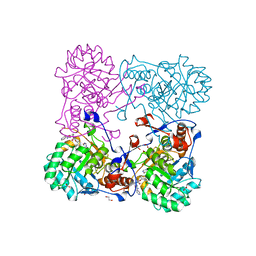 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor Q21 | | Descriptor: | (2S)-2-(naphthalen-1-yloxy)-N-[2-(pyridin-4-yl)-1,3-benzoxazol-5-yl]propanamide, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Kavitha, M, Cuny, G, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2924 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor Q21
To be Published, 2013
|
|
4MZ8
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with an Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5004 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91
To be Published
|
|
3N99
 
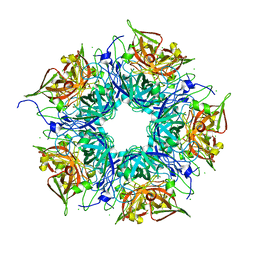 | | Crystal structure of TM1086 | | Descriptor: | CHLORIDE ION, uncharacterized protein TM1086 | | Authors: | Chruszcz, M, Domagalski, M.J, Wang, S, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-28 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of TM1086
To be Published
|
|
4PIB
 
 | | Crystal Structure of Uncharacterized Conserved Protein PixA from Burkholderia thailandensis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-05-08 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Uncharacterized Conserved Protein PixA from Burkholderia thailandensis
To Be Published
|
|
4PDY
 
 | | Crystal structure of aminoglycoside phosphotransferase from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | Aminoglycoside phosphotransferase, HISTIDINE, THIOCYANATE ION | | Authors: | Chang, C, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-22 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of aminoglycoside phosphotransferase from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446
To be published
|
|
2OGG
 
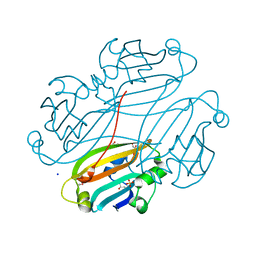 | | Structure of B. subtilis trehalose repressor (TreR) effector binding domain | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Rezacova, P, Krejcirikova, V, Borek, D, Moy, S.F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the effector-binding domain of the trehalose repressor TreR from Bacillus subtilis 168 reveals a unique quarternary assembly.
Proteins, 69, 2007
|
|
4Q6J
 
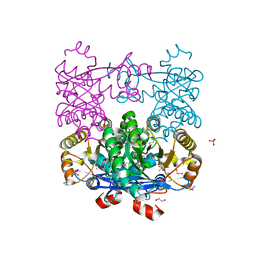 | | Crystal Structure of EAL domain Protein from Listeria monocytogenes EGD-e | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-22 | | Release date: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (1.369 Å) | | Cite: | Crystal Structure of EAL domain Protein from Listeria monocytogenes EGD-e
To be Published
|
|
2OKU
 
 | |
3NA2
 
 | | Crystal Structure of Protein of Unknown Function from Mine Drainage Metagenome Leptospirillum rubarum | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Uncharacterized protein | | Authors: | Kim, Y, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-31 | | Release date: | 2010-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Crsystal Structure of Protein of Unknown Function from Mine Drainage Metagenome Leptospirillum rubarum
To be Published
|
|
4PWT
 
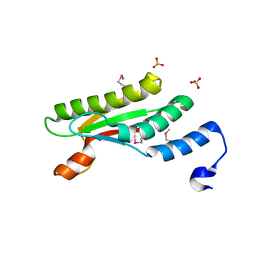 | | Crystal structure of peptidoglycan-associated outer membrane lipoprotein from Yersinia pestis CO92 | | Descriptor: | FORMIC ACID, PYROPHOSPHATE 2-, Peptidoglycan-associated lipoprotein, ... | | Authors: | Maltseva, N, Kim, Y, Osipiuk, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Crystal structure of peptidoglycan-associated outer membrane lipoprotein from
Yersinia pestis CO92
To be Published
|
|
4PZJ
 
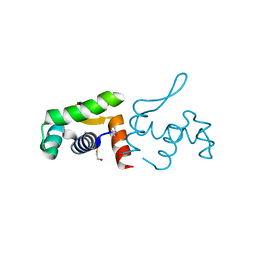 | | 1.60 Angstrom resolution crystal structure of a transcriptional regulator of the LysR family from Eggerthella lenta DSM 2243 | | Descriptor: | CHLORIDE ION, Transcriptional regulator, LysR family | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Kiryukhina, O, Endres, M, Shuvalova, L, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-31 | | Release date: | 2014-04-23 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.60 Angstrom resolution crystal structure of a transcriptional regulator of the LysR family from Eggerthella lenta DSM 2243
To be Published
|
|
2O2A
 
 | |
4PW0
 
 | |
2O3G
 
 | | Structural Genomics, the crystal structure of a conserved putative domain from Neisseria meningitidis MC58 | | Descriptor: | 1,2-ETHANEDIOL, Putative protein | | Authors: | Tan, K, Volkart, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of a conserved putative domain from Neisseria meningitidis MC58
To be Published
|
|
2OCD
 
 | | Crystal structure of L-asparaginase I from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | ACETATE ION, GLYCEROL, L-asparaginase I | | Authors: | Nocek, B, Wu, R, Osipiuk, J, Moy, S, Kim, Y, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-20 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of L-asparaginase I from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
4Q62
 
 | | Crystal Structure of Leucine-rich repeat- and Coiled coil-containing Protein from Legionella pneumophila | | Descriptor: | 1,2-ETHANEDIOL, Leucine-rich repeat-and coiled coil-containing protein, SULFATE ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2014-04-20 | | Release date: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal Structure of Leucine-rich repeat- and Coiled coil-containing Protein from
Legionella pneumophila
To be Published
|
|
2GAX
 
 | | Structure of Protein of Unknown Function Atu0240 from Agrobacteriium tumerfaciencs str. C58 | | Descriptor: | PHOSPHATE ION, hypothetical protein Atu0240 | | Authors: | Binkowski, T.A, Evdokimova, E, Kudritska, M, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-09 | | Release date: | 2006-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Hypothetical protein Atu0240 from Agrobacteriium tumerfaciencs str. C58
TO BE PUBLISHED
|
|
4Q7O
 
 | | The crystal structure of an immunity protein NMB0503 from Neisseria meningitidis MC58 | | Descriptor: | BROMIDE ION, FORMIC ACID, Immunity protein | | Authors: | Tan, K, Stols, L, Eschenfeldt, W, Babnigg, G, Low, D.A, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of a contact-dependent growth-inhibition (CDI) immunity protein from Neisseria meningitidis MC58.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
3IWF
 
 | | The Crystal Structure of the N-terminal domain of a RpiR Transcriptional Regulator from Staphylococcus epidermidis to 1.4A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-METHOXYETHANOL, CHLORIDE ION, ... | | Authors: | Stein, A.J, Sather, A, Borovilos, M, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-15 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the N-terminal domain of a RpiR Transcriptional Regulator from Staphylococcus epidermidis to 1.4A
To be Published
|
|
4MTN
 
 | |
4N03
 
 | | Fatty acid ABC transporter substrate-binding protein from Thermomonospora curvata | | Descriptor: | 1,2-ETHANEDIOL, ABC-type branched-chain amino acid transport systems periplasmic component-like protein, PALMITIC ACID | | Authors: | Osipiuk, J, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fatty acid ABC transporter substrate-binding protein from Thermomonospora curvata
To be Published
|
|
4MY9
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor C91 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, MALONATE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5893 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor C91
To be Published
|
|
2O3F
 
 | | Structural Genomics, the crystal structure of the N-terminal domain of the putative transcriptional regulator ybbH from Bacillus subtilis subsp. subtilis str. 168. | | Descriptor: | Putative HTH-type transcriptional regulator ybbH, SULFATE ION | | Authors: | Tan, K, Bigelow, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the N-terminal domain of the putative transcriptional regulator ybbH from Bacillus subtilis subsp. subtilis str. 168.
To be Published
|
|
3F3K
 
 | | The structure of uncharacterized protein YKR043C from Saccharomyces cerevisiae. | | Descriptor: | GLYCEROL, Uncharacterized protein YKR043C | | Authors: | Cuff, M, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae.
J.Biol.Chem., 285, 2010
|
|
3FRM
 
 | | The crystal structure of a functionally unknown conserved protein from Staphylococcus epidermidis ATCC 12228. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SODIUM ION, SULFATE ION, ... | | Authors: | Tan, K, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-08 | | Release date: | 2009-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein from Staphylococcus epidermidis ATCC 12228.
To be Published
|
|
