6TSP
 
 | | Marasmius oreades agglutinin (MOA) inhibited by zinc | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin, CALCIUM ION, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2019-12-21 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of MOA in complex with a peptide fragment: a protease caught in flagranti
Curr Res Struct Biol, 2, 2020
|
|
3F9R
 
 | | Crystal Structure of Trypanosoma Brucei phosphomannosemutase, TB.10.700.370 | | Descriptor: | MAGNESIUM ION, Phosphomannomutase, SULFATE ION | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Lin, Y.H, Guther, L, Shamshad, A, MacKenzie, F, Bandini, G, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-14 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Trypanosoma Brucei phosphomannosemutase, TB.10.700.370
To be Published
|
|
3ZTF
 
 | | X-ray Structure of the Cyan Fluorescent Protein mTurquoise2 (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-07-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
1GKG
 
 | | Structure Determination and Rational Mutagenesis reveal binding surface of immune adherence receptor, CR1 (CD35) | | Descriptor: | COMPLEMENT RECEPTOR TYPE 1 | | Authors: | Smith, B.O, Mallin, R.L, Krych-Goldberg, M, Wang, X, Hauhart, R.E, Bromek, K, Uhrin, D, Atkinson, J.P, Barlow, P.N. | | Deposit date: | 2001-08-14 | | Release date: | 2002-04-18 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the C3B Binding Site of Cr1 (Cd35), the Immune Adherence Receptor
Cell(Cambridge,Mass.), 108, 2002
|
|
6TUB
 
 | | Beta-endorphin amyloid fibril | | Descriptor: | Beta-endorphin | | Authors: | Verasdonck, J, Seuring, C, Gath, J, Ghosh, D, Nespovitaya, N, Waelti, M.A, Maji, S, Cadalbert, R, Boeckmann, A, Guentert, P, Meier, B.H, Riek, R. | | Deposit date: | 2020-01-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The three-dimensional structure of human beta-endorphin amyloid fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8B9F
 
 | | Structure of Echovirus 11 complexed with DAF (CD55) calculated from symmetry expansion | | Descriptor: | Complement decay-accelerating factor, Genome polyprotein, SPHINGOSINE | | Authors: | Stuart, D.I, Ren, J, Qin, L, Zhou, D. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-07 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Switching of Receptor Binding Poses between Closely Related Enteroviruses.
Viruses, 14, 2022
|
|
1FZD
 
 | | STRUCTURE OF RECOMBINANT ALPHAEC DOMAIN FROM HUMAN FIBRINOGEN-420 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Spraggon, G, Applegate, D, Everse, S.J, Zhang, J.-Z, Veerapandian, L, Redman, C, Doolittle, R.F, Grieninger, G. | | Deposit date: | 1998-06-22 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a recombinant alphaEC domain from human fibrinogen-420.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2OGG
 
 | | Structure of B. subtilis trehalose repressor (TreR) effector binding domain | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Rezacova, P, Krejcirikova, V, Borek, D, Moy, S.F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the effector-binding domain of the trehalose repressor TreR from Bacillus subtilis 168 reveals a unique quarternary assembly.
Proteins, 69, 2007
|
|
3J9Q
 
 | | Atomic structures of a bactericidal contractile nanotube in its pre- and post-contraction states | | Descriptor: | sheath, tube | | Authors: | Ge, P, Scholl, D, Leiman, P.G, Yu, X, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3ZOS
 
 | | Structure of the DDR1 kinase domain in complex with ponatinib | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Bradley, A, Coutandin, D, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-02-22 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Mechanisms Determining Inhibition of the Collagen Receptor Ddr1 by Selective and Multi-Targeted Type II Kinase Inhibitors
J.Mol.Biol., 426, 2014
|
|
3K8T
 
 | | Structure of eukaryotic rnr large subunit R1 complexed with designed adp analog compound | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2'-deoxy-2'-(2-hydroxyethyl)adenosine 5'-(trihydrogen diphosphate), GLYCEROL, ... | | Authors: | Sun, D, Xu, H, Dealwis, C, Lee, R.E. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design, Synthesis, and Evaluation of 2'-(2-Hydroxyethyl)-2'-deoxyadenosine and the 5'-Diphosphate Derivative as Ribonucleotide Reductase Inhibitors
Chemmedchem, 4, 2009
|
|
2OK6
 
 | |
3ZUZ
 
 | | Structure of Shq1p C-terminal domain | | Descriptor: | ISOPROPYL ALCOHOL, PROTEIN SHQ1 | | Authors: | Walbott, H, Machado-Pinilla, R, Liger, D, Blaud, M, Rety, S, Grozdanov, P.N, Godin, K, vanTilbeurgh, H, Varani, G, Meier, U.T, Leulliot, N. | | Deposit date: | 2011-07-22 | | Release date: | 2011-11-30 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The H/Aca Rnp Assembly Factor Shq1 Functions as an RNA Mimic.
Genes Dev., 25, 2011
|
|
6T5J
 
 | | Structure of NUDT15 in complex with inhibitor TH1760 | | Descriptor: | 6-[4-(1~{H}-indol-5-ylcarbonyl)piperazin-1-yl]sulfonyl-3~{H}-1,3-benzoxazol-2-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Carter, M, Rehling, D, Desroses, M, Zhang, S.M, Hagenkort, A, Valerie, N.C.K, Helleday, T, Stenmark, P. | | Deposit date: | 2019-10-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a chemical probe against NUDT15.
Nat.Chem.Biol., 16, 2020
|
|
8BBF
 
 | | Structure of the IFT-A complex; IFT-A1 module | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, WD repeat-containing protein 19 | | Authors: | Hesketh, S.J, Mukhopadhyay, A.G, Nakamura, D, Toropova, K, Roberts, A.J. | | Deposit date: | 2022-10-12 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | IFT-A structure reveals carriages for membrane protein transport into cilia.
Cell, 185, 2022
|
|
3K94
 
 | | Crystal Structure of Thiamin pyrophosphokinase from Geobacillus thermodenitrificans, Northeast Structural Genomics Consortium Target GtR2 | | Descriptor: | Thiamin pyrophosphokinase | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Janjua, J, Xiao, R, Patel, D.J, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Northeast Structural Genomics Consortium Target GtR2
To be Published
|
|
8BSD
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with tubercidin | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, ... | | Authors: | Kremling, V, Oberthuer, D, Sprenger, J. | | Deposit date: | 2022-11-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of Tubercidin bound to the active site of the SARS-CoV-2 methyltransferase nsp10-16
To Be Published
|
|
2NWI
 
 | | Crystal structure of protein AF1396 from Archaeoglobus fulgidus, Pfam DUF98 | | Descriptor: | Hypothetical protein | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Adams, J, Sridhar, V, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of hypothetical protein O28875 from Archaeoglobus fulgidus
To be Published
|
|
8B8R
 
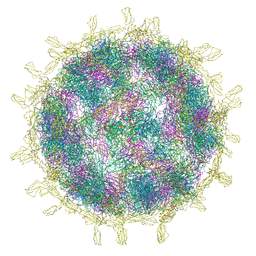 | | Complex of Echovirus 11 with its attaching receptor decay-accelerating factor (CD55) | | Descriptor: | DECAY ACCELERATING FACTOR (CD55), SPHINGOSINE, VP1, ... | | Authors: | Stuart, D.I, Ren, J, Zhou, D, Qin, L. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-07 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Switching of Receptor Binding Poses between Closely Related Enteroviruses.
Viruses, 14, 2022
|
|
3ZV7
 
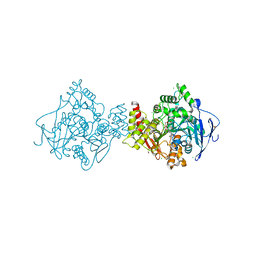 | | Torpedo californica Acetylcholinesterase Inhibition by Bisnorcymserine | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bartolucci, C, Stojan, J, Greig, N.H, Lamba, D. | | Deposit date: | 2011-07-24 | | Release date: | 2012-05-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Kinetics of Torpedo Californica Acetylcholinesterase Inhibition by Bisnorcymserine and Crystal Structure of the Complex with its Leaving Group.
Biochem.J., 444, 2012
|
|
3J1Z
 
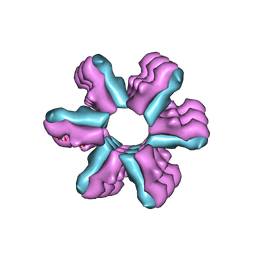 | | Inward-Facing Conformation of the Zinc Transporter YiiP revealed by Cryo-electron Microscopy | | Descriptor: | Cation efflux family protein | | Authors: | Coudray, N, Valvo, S, Hu, M, Lasala, R, Kim, C, Vink, M, Zhou, M, Provasi, D, Filizola, M, Tao, J, Fang, J, Penczek, P.A, Ubarretxena-Belandia, I, Stokes, D.L, Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS) | | Deposit date: | 2012-07-24 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Inward-facing conformation of the zinc transporter YiiP revealed by cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3J7H
 
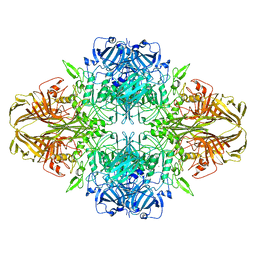 | | Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy | | Descriptor: | Beta-galactosidase, MAGNESIUM ION | | Authors: | Bartesaghi, A, Matthies, D, Banerjee, S, Merk, A, Subramaniam, S. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of beta-galactosidase at 3.2- angstrom resolution obtained by cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1G87
 
 | | THE CRYSTAL STRUCTURE OF ENDOGLUCANASE 9G FROM CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOCELLULASE 9G, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.P, Aghajari, N, Driguez, H, Haser, R. | | Deposit date: | 2000-11-16 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-Ray Crystal Structure of the Multidomain Endoglucanase Cel9G from Clostridium
cellulolyticum Complexed with Natural and Synthetic Cello-Oligosaccharides
J.BACTERIOL., 185, 2003
|
|
1G8Q
 
 | | CRYSTAL STRUCTURE OF HUMAN CD81 EXTRACELLULAR DOMAIN, A RECEPTOR FOR HEPATITIS C VIRUS | | Descriptor: | CD81 ANTIGEN, EXTRACELLULAR DOMAIN | | Authors: | Kitadokoro, K, Bolognesi, M, Bordo, D, Grandi, G, Galli, G, Petracca, R, Falugi, F. | | Deposit date: | 2000-11-20 | | Release date: | 2001-02-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs.
EMBO J., 20, 2001
|
|
3DXN
 
 | | Crystal structure of the calcium-dependent kinase from toxoplasma gondii, 541.m00134, kinase domain. | | Descriptor: | Calmodulin-like domain protein kinase isoform 3 | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Cossar, D, Wasney, G, Lin, Y.H, Hassani, A, Ali, A, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Wikstrom, M, Edwards, A.M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-24 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of the calcium-dependent kinase from toxoplasma gondii, 541.m00134, kinase domain.
To be Published
|
|
