7DSQ
 
 | | Overall structure of the LAT1-4F2hc bound with 3,5-diiodo-L-tyrosine | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,5-DIIODOTYROSINE, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Zhong, X.Y, Zhou, Q. | | Deposit date: | 2020-12-31 | | Release date: | 2021-03-10 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of substrate transport and inhibition of the human LAT1-4F2hc amino acid transporter.
Cell Discov, 7, 2021
|
|
7DSN
 
 | | Overall structure of the LAT1-4F2hc bound with JX-119 | | Descriptor: | (2~{S})-2-azanyl-7-[[2-(1,3-benzoxazol-2-yl)phenyl]methoxy]-3,4-dihydro-1~{H}-naphthalene-2-carboxylic acid, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Zhong, X.Y, Zhou, Q. | | Deposit date: | 2020-12-31 | | Release date: | 2021-03-10 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of substrate transport and inhibition of the human LAT1-4F2hc amino acid transporter.
Cell Discov, 7, 2021
|
|
7DSK
 
 | | Overall structure of the LAT1-4F2hc bound with JX-075 | | Descriptor: | (2~{S})-2-azanyl-7-(naphthalen-1-ylmethoxy)-3,4-dihydro-1~{H}-naphthalene-2-carboxylic acid, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Zhong, X.Y, Zhou, Q. | | Deposit date: | 2020-12-31 | | Release date: | 2021-03-10 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of substrate transport and inhibition of the human LAT1-4F2hc amino acid transporter.
Cell Discov, 7, 2021
|
|
8QNZ
 
 | | Crystal Structure of a Class D Carbapenemase Complexed with Hydrolyzed Imipenem | | Descriptor: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, 1-BUTANOL, BROMIDE ION, ... | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2023-09-27 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity.
Acs Cent.Sci., 9, 2023
|
|
3JCT
 
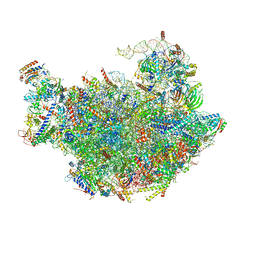 | | Cryo-em structure of eukaryotic pre-60S ribosomal subunits | | Descriptor: | 60S ribosomal protein L11-A, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Wu, S, Kumcuoglu, B, Yan, K.G, Brown, H, Zhang, Y.X, Tan, D, Gamalinda, M, Yuan, Y, Li, Z.F, Jakovljevic, J, Ma, C.Y, Lei, J.L, Dong, M.Q, Woolford Jr, J.L, Gao, N. | | Deposit date: | 2016-03-09 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes
Nature, 534, 2016
|
|
7PGO
 
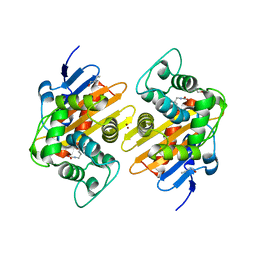 | | Crystal Structure of a Class D Carbapenemase_R250A | | Descriptor: | 1-BUTANOL, BROMIDE ION, Beta-lactamase | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2021-08-15 | | Release date: | 2022-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
5D6P
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[4-(hydroxymethyl)-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl]urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5D6Q
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-{4-[(E)-2-(pyridin-3-yl)ethenyl]-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl}urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5C2Y
 
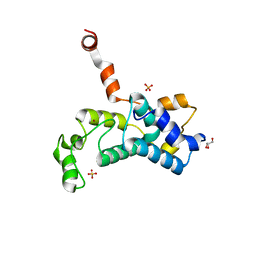 | | Crystal structure of the Saccharomyces cerevisiae Rtr1 (regulator of transcription) | | Descriptor: | GLYCEROL, RNA polymerase II subunit B1 CTD phosphatase RTR1, SULFATE ION, ... | | Authors: | Yogesha, S.D, Irani, S, Zhang, Y.J. | | Deposit date: | 2015-06-16 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Saccharomyces cerevisiae Rtr1 reveals an active site for an atypical phosphatase.
Sci.Signal., 9, 2016
|
|
5D7C
 
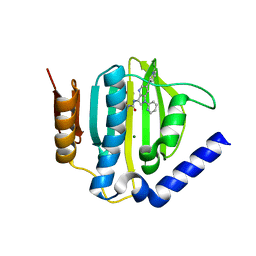 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[1-(pyridin-2-yl)-6-(pyridin-3-yl)-1H-pyrrolo[3,2-b]pyridin-3-yl]urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
1P9G
 
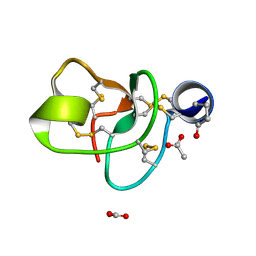 | | Crystal structure of a novel antifungal protein distinct with five disulfide bridges from Ecommia ulmoides Oliver at atomic resolution | | Descriptor: | ACETATE ION, EAFP 2 | | Authors: | Xiang, Y, Huang, R.H, Liu, X.Z, Wang, D.C. | | Deposit date: | 2003-05-12 | | Release date: | 2004-06-01 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Crystal structure of a novel antifungal protein distinct with five disulfide bridges from Eucommia ulmoides Oliver at an atomic resolution.
J.Struct.Biol., 148, 2004
|
|
5J8V
 
 | |
8Z99
 
 | |
8YHD
 
 | |
8Z9C
 
 | |
8Z4L
 
 | |
8YHE
 
 | |
8Z4J
 
 | |
8Z9E
 
 | |
5LG4
 
 | |
8HIF
 
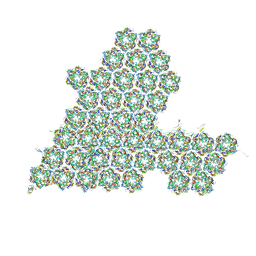 | | One asymmetric unit of Singapore grouper iridovirus capsid | | Descriptor: | Major capsid protein, Penton protein (VP14), VP137, ... | | Authors: | Zhao, Z.N, Liu, C.C, Zhu, D.J, Qi, J.X, Zhang, X.Z, Gao, G.F. | | Deposit date: | 2022-11-20 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Near-atomic architecture of Singapore grouper iridovirus and implications for giant virus assembly.
Nat Commun, 14, 2023
|
|
4FF5
 
 | |
7LRS
 
 | |
5M4Y
 
 | |
1FPL
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE (D-FRUCTOSE-1,6-BISPHOSPHATE 1-PHOSPHOHYDROLASE) COMPLEXED WITH AMP, 2,5-ANHYDRO-D-GLUCITOL-1,6-BISPHOSPHATE AND THALLIUM IONS (10 MM) | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Villeret, V, Lipscomb, W.N. | | Deposit date: | 1995-06-02 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic evidence for the action of potassium, thallium, and lithium ions on fructose-1,6-bisphosphatase.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
