9LBN
 
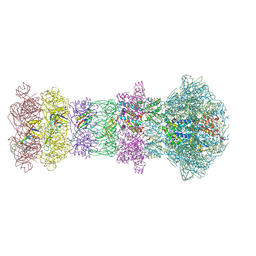 | | The composite cryo-EM structure of the head-to-tail connector and head-proximal tail components of bacteriophage phiXacJX1 | | Descriptor: | adaptor protein gp5, portal protein gp1, stopper protein gp6, ... | | Authors: | Guo, M, Wang, A, Zheng, Y, Liu, C, Shao, Q, Fang, Q. | | Deposit date: | 2025-01-03 | | Release date: | 2025-05-07 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of a Xanthomonas phage: Insights into viral architecture and implications for the model phage HK97.
Structure, 33, 2025
|
|
9MOZ
 
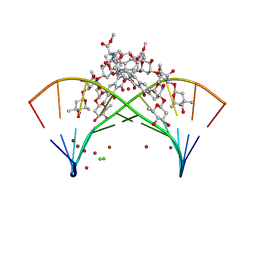 | | Crystal structure of mithramycin analogue MTM SA-5-methyl-Trp in complex with DNA AGAGGCCTCT | | Descriptor: | CHLORIDE ION, DNA (5'-D(*AP*GP*AP*GP*GP*CP*CP*TP*CP*T)-3'), SODIUM ION, ... | | Authors: | Hou, C, Tsodikov, O.V. | | Deposit date: | 2024-12-28 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The Position of Indole Methylation Controls the Structure, DNA Binding, and Cellular Functions of Mithramycin SA-Trp Analogues.
Chembiochem, 26, 2025
|
|
9MR6
 
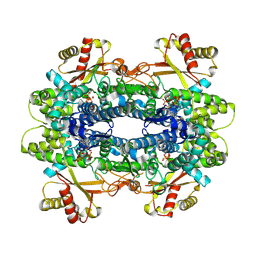 | | X-ray crystal structure of SAMHD1 from Rhizophagus irregularis | | Descriptor: | CALCIUM ION, GUANOSINE-5'-TRIPHOSPHATE, HD domain-containing protein, ... | | Authors: | Lachowicz, J.C, Zizola, C, Grove, T.G. | | Deposit date: | 2025-01-07 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Activation and Allostery in a Fungal SAMHD1 Hydrolase: An Evolutionary Blueprint for dNTP Catabolism.
Jacs Au, 5, 2025
|
|
8RBN
 
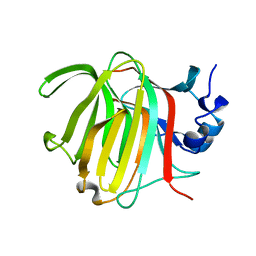 | |
9OMF
 
 | | Cryo-EM structure of neddylated PCMTD1-ELOBC-CUL5-RBX2 (N8-CRL5-PCMTD1) | | Descriptor: | Cullin-5, Elongin-B, Elongin-C, ... | | Authors: | Pang, E.Z, Zhao, B, Flowers, C, Oroudjeva, E, Winters, J.B, Pandey, V, Sawaya, M.R, Wohlschlegel, W, Loo, J.A, Rodriguez, J.A, Clarke, S.G. | | Deposit date: | 2025-05-13 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (9.72 Å) | | Cite: | Structural basis for L-isoaspartyl-containing protein recognition by the PCMTD1 cullin-RING E3 ubiquitin ligase.
Biorxiv, 2025
|
|
9QLS
 
 | |
9NEV
 
 | | Acanthamoeba Polyphaga Mimivirus L230 | | Descriptor: | Procollagen lysyl hydroxylase and glycosyltransferase | | Authors: | Chen, T, Buhlheller, C, Guo, H. | | Deposit date: | 2025-02-20 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of an Fe 2+ -binding-deficient mimiviral collagen lysyl hydroxylase.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
9V6Z
 
 | | Crystal structure of Isoform Chitin Binding Protein from Iberis umbellata L. | | Descriptor: | ACETATE ION, CHLORIDE ION, Chitin Binding Protein III, ... | | Authors: | Saeed, A, Betzel, C, Brognaro, H, Rajaiah Prabhu, P, Alves Franca, B, Mehmood, S, Khaliq, B, Ishaq, U, Akrem, A. | | Deposit date: | 2025-05-27 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure of Isoform Chitin Binding Protein from Iberis umbellata L.
To Be Published
|
|
9LON
 
 | | Cryo-EM structure of alpha-synuclein P8 fibril | | Descriptor: | Alpha-synuclein | | Authors: | Xia, W.C, Liu, C. | | Deposit date: | 2025-01-23 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Fibril fuzzy coat is important for alpha-synuclein pathological transmission activity.
Neuron, 113, 2025
|
|
8RBR
 
 | |
9MEX
 
 | | Structure of phosphocysteine intermediate of human PRL1 phosphatase | | Descriptor: | Protein tyrosine phosphatase type IVA 1, SULFATE ION | | Authors: | Mahbub, L, Kozlov, G, Knorn, C, Gehring, K. | | Deposit date: | 2024-12-09 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the phosphocysteine intermediate of the phosphatase of regenerating liver PTP4A1.
J.Biol.Chem., 301, 2025
|
|
9QKV
 
 | |
8RE3
 
 | | Crystal Structure determination of Dye-decolorizing Peroxidase (DyP) mutant M190G from Deinoccoccus radiodurans | | Descriptor: | CALCIUM ION, CHLORIDE ION, HYDROXIDE ION, ... | | Authors: | Salgueiro, B.A, Frade, K, Frazao, C, Matias, P, Moe, E. | | Deposit date: | 2023-12-10 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biochemical, Biophysical, and Structural Analysis of an Unusual DyP from the Extremophile Deinococcus radiodurans.
Molecules, 29, 2024
|
|
9QNV
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-5-phenyl-[1,3]thiazolo[2,3-c][1,2,4]triazole, Bromodomain-containing protein 4, ... | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-03-25 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.226 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound 2
To Be Published
|
|
8RE2
 
 | | Crystal Structure determination of Dye-decolorizing Peroxidase (DyP) from Deinoccoccus radiodurans | | Descriptor: | GLYCEROL, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Salgueiro, B.A, Frade, K, Frazao, C, Moe, E. | | Deposit date: | 2023-12-10 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical, Biophysical, and Structural Analysis of an Unusual DyP from the Extremophile Deinococcus radiodurans.
Molecules, 29, 2024
|
|
9UWD
 
 | | Cryo-EM structure of inactive-DP1 | | Descriptor: | Prostaglandin D2 receptor,Soluble cytochrome b562 | | Authors: | Xu, J, Xu, Y, Wu, C, Xu, H.E. | | Deposit date: | 2025-05-12 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular basis for ligand recognition and receptor activation of the prostaglandin D2 receptor DP1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8RI9
 
 | | Late alpha-Synuclein fibril structure from liquid-liquid phase separations. | | Descriptor: | Alpha-synuclein | | Authors: | De Simone, A, Barritt, J.D, Chen, S, Cascella, R, Cecchi, C, Bigi, A, Jarvis, J.A, Chiti, F, Dobson, C.M, Fusco, G. | | Deposit date: | 2023-12-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-Toxicity Relationship in Intermediate Fibrils from alpha-Synuclein Condensates.
J.Am.Chem.Soc., 146, 2024
|
|
8S5N
 
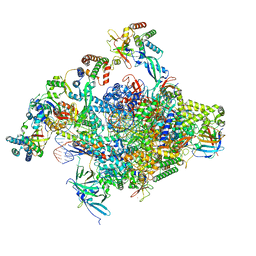 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 12 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-24 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8RCW
 
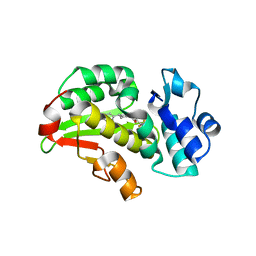 | | Crystal structure of the Mycobacterium tuberculosis regulator VirS (N-terminal fragment 4-208) in complex with the lead compound SMARt751 | | Descriptor: | 4,4,4-tris(fluoranyl)-1-[4-(4-fluorophenyl)piperidin-1-yl]butan-1-one, HTH-type transcriptional regulator VirS | | Authors: | Grosse, C, Sigoillot, M, Megalizzi, V, Tanina, A, Willand, N, Baulard, A.R, Wintjens, R. | | Deposit date: | 2023-12-07 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis VirS regulator reveals its interaction with the lead compound SMARt751.
J.Struct.Biol., 216, 2024
|
|
8RBO
 
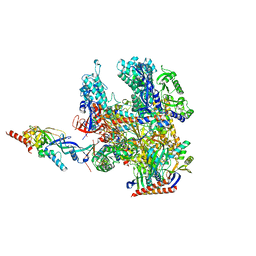 | | Cryo-EM structure of Pyrococcus furiosus apo form RNA polymerase contracted clamp conformation | | Descriptor: | DNA-directed RNA polymerase subunit Rpo10, DNA-directed RNA polymerase subunit Rpo11, DNA-directed RNA polymerase subunit Rpo12, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-12-04 | | Release date: | 2024-04-24 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
8RQP
 
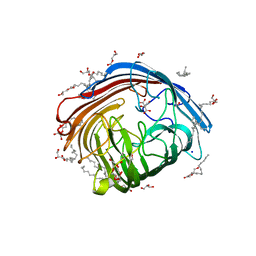 | | In meso structure of the alginate exporter, AlgE, from Pseudomonas aeruginosa in 7.10 monoacylglycerol | | Descriptor: | 7.10 monoacylglycerol (R-form), 7.10 monoacylglycerol (S-form), Alginate production protein AlgE, ... | | Authors: | Smithers, L, Boland, C, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
8RQR
 
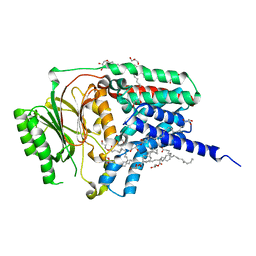 | | In meso structure of apolipoprotein N-acyltransferase, Lnt, from Escherichia coli in 7.10 monoacylglycerol | | Descriptor: | 7.10 monoacylglycerol (S-form), Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Smithers, L, Boland, C, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
8S55
 
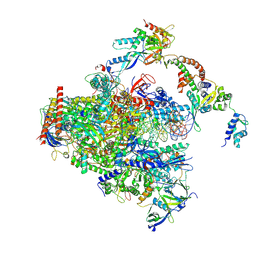 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state a (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S51
 
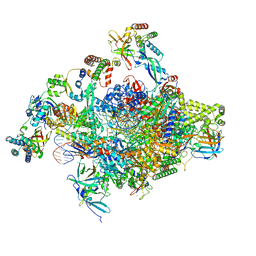 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 8 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8RM6
 
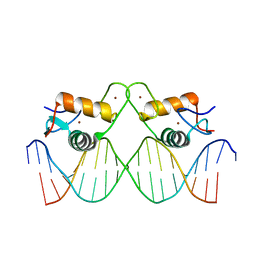 | | Crystal Structure of Human Androgen Receptor DNA Binding Domain Bound to its Response Element: C3(1)ARE | | Descriptor: | C3(1)ARE_Chain C, C3(1)ARE_Chain D, Isoform 2 of Androgen receptor, ... | | Authors: | Lee, X.Y, Helsen, C, Van Eynde, W, Voet, A, Claessens, F. | | Deposit date: | 2024-01-05 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural mechanism underlying variations in DNA binding by the androgen receptor.
J.Steroid Biochem.Mol.Biol., 241, 2024
|
|
