8DMO
 
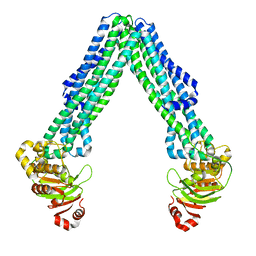 | | Structure of open, inward-facing MsbA from E. coli | | Descriptor: | ATP-binding transport protein MsbA | | Authors: | Liu, C, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2022-07-08 | | Release date: | 2022-12-14 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for lipid and copper regulation of the ABC transporter MsbA.
Nat Commun, 13, 2022
|
|
8DMM
 
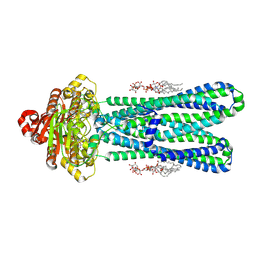 | | Structure of the vanadate-trapped MsbA bound to KDL | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-5-oxidanyl-oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ADP ORTHOVANADATE, ATP-binding transport protein MsbA | | Authors: | Liu, C, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2022-07-08 | | Release date: | 2022-12-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis for lipid and copper regulation of the ABC transporter MsbA.
Nat Commun, 13, 2022
|
|
8DZJ
 
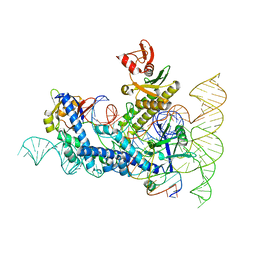 | |
8TSR
 
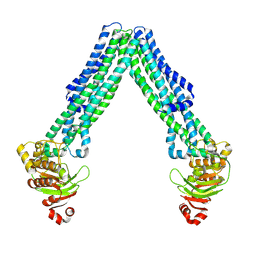 | | Open, inward-facing MsbA structure (OIF4) | | Descriptor: | ATP-binding transport protein MsbA | | Authors: | Yang, B, Zhang, T, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2023-08-11 | | Release date: | 2024-06-19 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Native mass spectrometry and structural studies reveal modulation of MsbA-nucleotide interactions by lipids.
Nat Commun, 15, 2024
|
|
8TSO
 
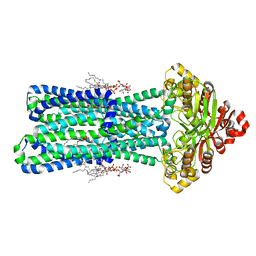 | | KDL bound, nucleotide-free MsbA in open, outward-facing conformation | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-5-oxidanyl-oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-binding transport protein MsbA, PENTAETHYLENE GLYCOL MONODECYL ETHER | | Authors: | Yang, B, Zhang, T, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2023-08-11 | | Release date: | 2024-06-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Native mass spectrometry and structural studies reveal modulation of MsbA-nucleotide interactions by lipids.
Nat Commun, 15, 2024
|
|
8TSP
 
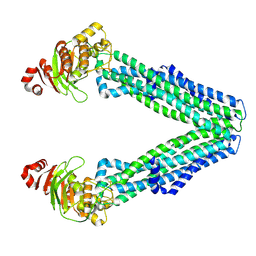 | | Open, inward-facing MsbA structure (OIF1) | | Descriptor: | ATP-binding transport protein MsbA | | Authors: | Yang, B, Zhang, T, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2023-08-11 | | Release date: | 2024-06-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Native mass spectrometry and structural studies reveal modulation of MsbA-nucleotide interactions by lipids.
Nat Commun, 15, 2024
|
|
8TSQ
 
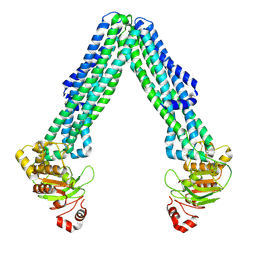 | | Open, inward-facing MsbA structure (OIF2) | | Descriptor: | ATP-binding transport protein MsbA | | Authors: | Yang, B, Zhang, T, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2023-08-11 | | Release date: | 2024-06-19 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Native mass spectrometry and structural studies reveal modulation of MsbA-nucleotide interactions by lipids.
Nat Commun, 15, 2024
|
|
8TSS
 
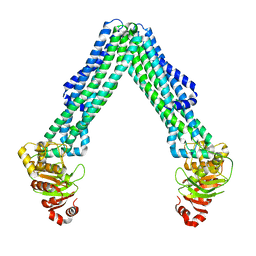 | | Open, inward-facing MsbA structure (OIF3) | | Descriptor: | ATP-binding transport protein MsbA | | Authors: | Yang, B, Zhang, T, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2023-08-11 | | Release date: | 2024-06-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Native mass spectrometry and structural studies reveal modulation of MsbA-nucleotide interactions by lipids.
Nat Commun, 15, 2024
|
|
1XE1
 
 | | Hypothetical Protein From Pyrococcus Furiosus Pfu-880080-001 | | Descriptor: | SULFATE ION, hypothetical protein PF0907 | | Authors: | Chang, J.C, Zhao, M, Zhou, W, Liu, Z.J, Tempel, W, Arendall III, W.B, Chen, L, Lee, D, Habel, J.E, Rose, J.P, Richardson, J.S, Richardson, D.C, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-08 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical Protein From Pyrococcus Furiosus Pfu-880080-001
To be published
|
|
1YCY
 
 | | Conserved hypothetical protein Pfu-1806301-001 from Pyrococcus furiosus | | Descriptor: | Conserved hypothetical protein | | Authors: | Huang, L, Liu, Z.-J, Lee, D, Tempel, W, Chang, J, Zhao, M, Habel, J, Xu, H, Chen, L, Nguyen, D, Chang, S.-H, Horanyi, P, Florence, Q, Zhou, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-23 | | Release date: | 2005-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conserved hypothetical protein Pfu-1806301-001 from Pyrococcus furiosus
To be published
|
|
1XMA
 
 | | Structure of a transcriptional regulator from Clostridium thermocellum Cth-833 | | Descriptor: | MERCURY (II) ION, Predicted transcriptional regulator, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Chen, L, Lee, D, Habel, J, Nguyen, J, Chang, S.-H, Kataeva, I, Xu, H, Chang, J, Zhao, M, Horanyi, P, Florence, Q, Zhou, W, Tempel, W, Lin, D, Praissman, J, Zhang, H, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Ljungdahl, L, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-10-01 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of a transcriptional regulator from Clostridium thermocellum Cth-833
To be published
|
|
1ZGH
 
 | | Methionyl-tRNA formyltransferase from Clostridium thermocellum | | Descriptor: | Methionyl-tRNA formyltransferase, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Kataeva, I, Xu, H, Zhao, M, Chang, J, Liu, Z, Chen, L, Tempel, W, Habel, J, Zhou, W, Lee, D, Lin, D, Chang, S, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Wang, B, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-04-21 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Methionyl-tRNA formyltransferase from Clostridium thermocellum
To be published
|
|
2ICU
 
 | | Crystal Structure of Hypothetical Protein YedK From Escherichia coli | | Descriptor: | Hypothetical protein yedK | | Authors: | Chen, L, Liu, Z.J, Li, Y, Zhao, M, Rose, J, Ebihara, A, Yokoyama, S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-13 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Hypothetical Protein YedK From Escherichia coli
To be Published
|
|
1YB3
 
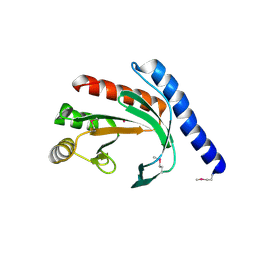 | | Conserved hypothetical protein Pfu-178653-001 from Pyrococcus furiosus | | Descriptor: | UNKNOWN ATOM OR ION, hypothetical protein | | Authors: | Habel, J, Zhou, W, Chang, J, Zhao, M, Xu, H, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Horanyi, P, Florence, Q, Tempel, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-19 | | Release date: | 2005-02-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conserved hypothetical protein Pfu-178653-001 from Pyrococcus furiosus
To be published
|
|
1YBX
 
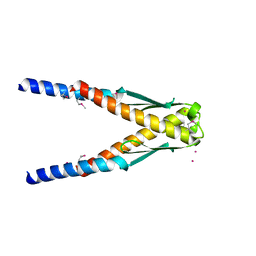 | | Conserved hypothetical protein Cth-383 from Clostridium thermocellum | | Descriptor: | Conserved hypothetical protein, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Chang, J, Zhao, M, Habel, J, Kataeva, I, Xu, H, Chen, L, Lee, D, Nguyen, J, Chang, S.-H, Horanyi, P, Florence, Q, Zhou, W, Lin, D, Zhang, H, Ljundahl, L, Liu, Z.-J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved hypothetical protein Cth-383 from Clostridium thermocellum
To be published
|
|
1YD7
 
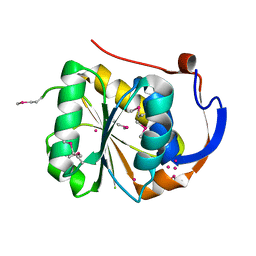 | | Conserved hypothetical protein Pfu-1647980-001 from Pyrococcus furiosus | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha, UNKNOWN ATOM OR ION | | Authors: | Horanyi, P, Florence, Q, Zhou, W, Chang, J, Zhao, M, Habel, J, Xu, H, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Tempel, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-23 | | Release date: | 2005-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved hypothetical protein Pfu-1647980-001 from Pyrococcus furiosus
To be published
|
|
2IEL
 
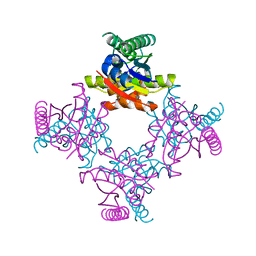 | | CRYSTAL STRUCTURE OF TT0030 from Thermus Thermophilus | | Descriptor: | Hypothetical Protein TT0030 | | Authors: | Zhu, J, Huang, J, Stepanyuk, G, Chen, L, Chang, J, Zhao, M, Xu, H, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-09-19 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF TT0030 from Thermus Thermophilus AT 1.6 ANGSTROMS RESOLUTION
To be Published
|
|
2HQE
 
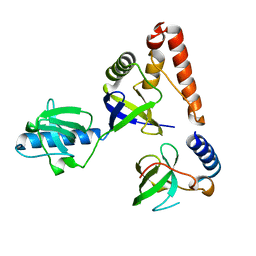 | | Crystal structure of human P100 Tudor domain: Large fragment | | Descriptor: | P100 Co-activator tudor domain | | Authors: | Shah, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Liu, Z.J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-18 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a large fragment of the Human P100 Tudor Domain
To be Published
|
|
1XI8
 
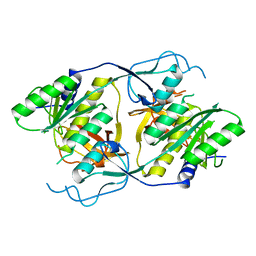 | | Molybdenum cofactor biosynthesis protein from Pyrococcus furiosus Pfu-1657500-001 | | Descriptor: | Molybdenum cofactor biosynthesis protein | | Authors: | Zhou, W, Zhao, M, Chang, J.C, Liu, Z.-J, Chen, L, Horanyi, P, Xu, H, Yang, H, Habel, J.E, Lee, D, Chang, S.-H, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Molybdenum cofactor biosynthesis protein from Pyrococcus furiosus Pfu-1657500-001
To be published
|
|
1XG7
 
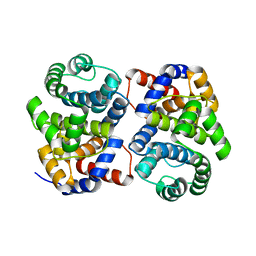 | | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Chang, J, Zhao, M, Horanyi, P, Xu, H, Yang, H, Liu, Z.-J, Chen, L, Zhou, W, Habel, J, Tempel, W, Lee, D, Lin, D, Chang, S.-H, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Chen, C.-Y, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus
To be published
|
|
2KQQ
 
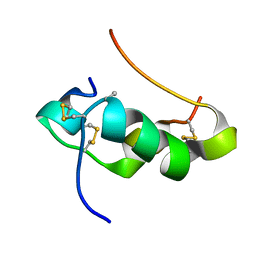 | | NMR structure of human insulin mutant gly-b8-d-ala, his-b10-asp, pro-b28-lys, lys-b29-pro, 20 structures | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Hua, Q.X, Wan, Z.L, Zhao, M, Jia, W, Huang, K, Weiss, M.A. | | Deposit date: | 2009-11-13 | | Release date: | 2010-11-24 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of an Turn-Stabilize But Ina Insulin Analog.
To be Published
|
|
4JHA
 
 | | Crystal Structure of RSV-Neutralizing Human Antibody D25 | | Descriptor: | D25 antigen-binding fragment heavy chain, D25 light chain | | Authors: | Mclellan, J.S, Chen, M, Leung, S, Graepel, K.W, Du, X, Yang, Y, Zhou, T, Baxa, U, Yasuda, E, Beaumont, T, Kumar, A, Modjarrad, K, Zheng, Z, Zhao, M, Xia, N, Kwong, P.D, Graham, B.S. | | Deposit date: | 2013-03-04 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody.
Science, 340, 2013
|
|
4JHW
 
 | | Crystal Structure of Respiratory Syncytial Virus Fusion Glycoprotein Stabilized in the Prefusion Conformation by Human Antibody D25 | | Descriptor: | D25 antigen-binding fragment heavy chain, D25 light chain, Fusion glycoprotein F0 | | Authors: | Mclellan, J.S, Chen, M, Leung, S, Graepel, K.W, Du, X, Yang, Y, Zhou, T, Baxa, U, Yasuda, E, Beaumont, T, Kumar, A, Modjarrad, K, Zheng, Z, Zhao, M, Xia, N, Kwong, P.D, Graham, B.S. | | Deposit date: | 2013-03-05 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody.
Science, 340, 2013
|
|
2O4X
 
 | | Crystal structure of human P100 tudor domain | | Descriptor: | Staphylococcal nuclease domain-containing protein 1 | | Authors: | Shaw, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Rao, Z, Wang, B.C, Liu, Z.J. | | Deposit date: | 2006-12-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human P100 tudor domain
To be Published
|
|
2P5U
 
 | | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase | | Authors: | Fu, Z.-Q, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Dillard, B, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD
To be Published
|
|
