5BTO
 
 | |
5BTB
 
 | | Crystal Structure of Ashbya gossypii Rai1 | | Descriptor: | AFR263Cp, SULFATE ION | | Authors: | Wang, V.Y, Tong, L. | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical studies of the distinct activity profiles of Rai1 enzymes.
Nucleic Acids Res., 43, 2015
|
|
5BTH
 
 | | Crystal structure of Candida albicans Rai1 | | Descriptor: | Decapping nuclease RAI1 | | Authors: | Wang, V.Y, Tong, L. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical studies of the distinct activity profiles of Rai1 enzymes.
Nucleic Acids Res., 43, 2015
|
|
5BUD
 
 | |
5CS4
 
 | |
1IEC
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT (H157A) OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
4MPS
 
 | | Crystal structure of rat Beta-galactoside alpha-2,6-sialyltransferase 1 (ST6GAL1), Northeast Structural Genomics Consortium Target RnR367A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactoside alpha-2,6-sialyltransferase 1 | | Authors: | Forouhar, F, Meng, L, Milaninia, S, Seetharaman, J, Su, M, Kornhaber, G, Montelione, G.T, Hunt, J.F, Moremen, K.W, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-09-13 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enzymatic Basis for N-Glycan Sialylation: STRUCTURE OF RAT alpha 2,6-SIALYLTRANSFERASE (ST6GAL1) REVEALS CONSERVED AND UNIQUE FEATURES FOR GLYCAN SIALYLATION.
J.Biol.Chem., 288, 2013
|
|
9DSO
 
 | |
9DUO
 
 | | Human PELP1-WDR18 complex | | Descriptor: | Proline-, glutamic acid- and leucine-rich protein 1, WD repeat-containing protein 18 | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2024-10-03 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Molecular insights into the overall architecture of human rixosome.
Nat Commun, 16, 2025
|
|
9DUN
 
 | | Human LAS1L-NOL9 complex | | Descriptor: | Polynucleotide 5'-hydroxyl-kinase NOL9, Ribosomal biogenesis protein LAS1L | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2024-10-03 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular insights into the overall architecture of human rixosome.
Nat Commun, 16, 2025
|
|
9DUM
 
 | | Human PELP1-WDR18-TEX10 complex | | Descriptor: | Proline-, glutamic acid- and leucine-rich protein 1, Testis-expressed protein 10, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2024-10-03 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Molecular insights into the overall architecture of human rixosome.
Nat Commun, 16, 2025
|
|
7K95
 
 | | Crystal structure of human CPSF30 in complex with hFip1 | | Descriptor: | Isoform 2 of Cleavage and polyadenylation specificity factor subunit 4, Pre-mRNA 3'-end-processing factor FIP1, ZINC ION | | Authors: | Hamilton, K, Tong, L. | | Deposit date: | 2020-09-28 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism for the interaction between human CPSF30 and hFip1.
Genes Dev., 34, 2020
|
|
2OA5
 
 | | Crystal structure of ORF52 from Murid herpesvirus (MUHV-4) (Murine gammaherpesvirus 68) at 2.1 A resolution. Northeast Structural Genomics Consortium target MHR28B. | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Hypothetical protein BQLF2 | | Authors: | Benach, J, Chen, Y, Seetharaman, J, Janjua, H, Xiao, R, Cunningham, K, Ma, L.-C, Ho, C.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-14 | | Release date: | 2007-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of the abundant tegument protein ORF52 from murine gammaherpesvirus 68.
J.Biol.Chem., 282, 2007
|
|
6APL
 
 | | Crystal Structure of human ST6GALNAC2 in complex with CMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Forouhar, F, Moremen, K.W, Northeast Structural Genomics Consortium (NESG), Tong, L. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Expression system for structural and functional studies of human glycosylation enzymes.
Nat. Chem. Biol., 14, 2018
|
|
6APJ
 
 | | Crystal Structure of human ST6GALNAC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 | | Authors: | Forouhar, F, Moremen, K.W, Northeast Structural Genomics Consortium (NESG), Tong, L. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Expression system for structural and functional studies of human glycosylation enzymes.
Nat. Chem. Biol., 14, 2018
|
|
3C96
 
 | | Crystal structure of the flavin-containing monooxygenase phzS from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium target PaR240 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Wang, D, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-15 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the flavin-containing monooxygenase phzS from Pseudomonas aeruginosa.
To be Published
|
|
8G1U
 
 | | Structure of the methylosome-Lsm10/11 complex | | Descriptor: | ADENOSINE, Methylosome protein 50, Methylosome subunit pICln, ... | | Authors: | Lin, M, Paige, A, Tong, L. | | Deposit date: | 2023-02-03 | | Release date: | 2023-08-23 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | In vitro methylation of the U7 snRNP subunits Lsm11 and SmE by the PRMT5/MEP50/pICln methylosome.
Rna, 29, 2023
|
|
8GXL
 
 | | HUMAN SUGP1 433-577 | | Descriptor: | SURP and G-patch domain-containing protein 1 | | Authors: | Xu, K, Tong, L. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DHX15 is involved in SUGP1-mediated RNA missplicing by mutant SF3B1 in cancer.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8GXM
 
 | | HUMAN SUGP1 433-586 | | Descriptor: | SURP and G-patch domain-containing protein 1 | | Authors: | Xu, K, Tong, L. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | DHX15 is involved in SUGP1-mediated RNA missplicing by mutant SF3B1 in cancer.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SN8
 
 | |
5HCU
 
 | |
7UG9
 
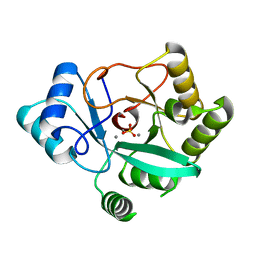 | | Crystal structure of RNase AM PHP domain | | Descriptor: | 5'-3' exoribonuclease, MANGANESE (II) ION, SULFATE ION | | Authors: | Doamekpor, S.K, Tong, L. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Identification of a novel deFADding activity in human, yeast and bacterial 5' to 3' exoribonucleases.
Nucleic Acids Res., 50, 2022
|
|
3TJV
 
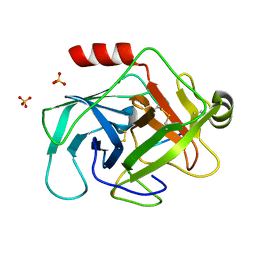 | | Crystal structure of human granzyme H with a peptidyl substrate | | Descriptor: | Granzyme H, PTSYAGDDSG, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
4LT9
 
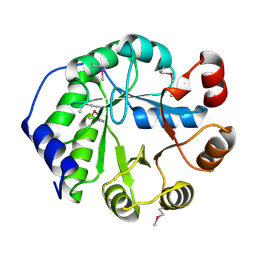 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR404 | | Descriptor: | Engineered Protein OR404 | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Sahdev, S, Xiao, R, Kogan, S, Maglaqui, M, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Engineered Protein OR404.
To be Published
|
|
3TJU
 
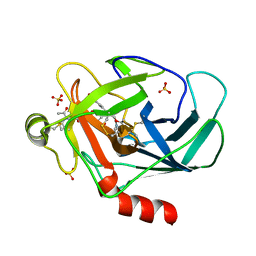 | | Crystal structure of human granzyme H with an inhibitor | | Descriptor: | Ac-PTSY-CMK inhibitor, Granzyme H, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
