4R5L
 
 | | Crystal structure of the DnaK C-terminus (Dnak-SBD-C) | | Descriptor: | CALCIUM ION, Chaperone protein DnaK, PHOSPHATE ION, ... | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9701 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
4R5I
 
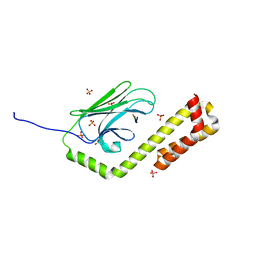 | | Crystal structure of the DnaK C-terminus with the substrate peptide NRLLLTG | | Descriptor: | Chaperone protein DnaK, HSP70/DnaK Substrate Peptide: NRLLLTG, PHOSPHATE ION, ... | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9702 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
4R5G
 
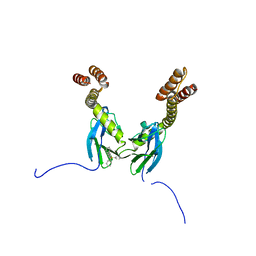 | | Crystal structure of the DnaK C-terminus with the inhibitor PET-16 | | Descriptor: | Chaperone protein DnaK, triphenyl(phenylethynyl)phosphonium | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4501 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
1W19
 
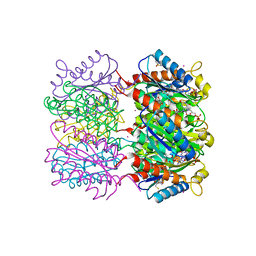 | | Lumazine Synthase from Mycobacterium tuberculosis bound to 3-(1,3,7- trihydro-9-D-ribityl-2,6,8-purinetrione-7-yl)propane 1-phosphate | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (4S,5S)-1,2-DITHIANE-4,5-DIOL, ... | | Authors: | Morgunova, E, Meining, W, Illarionov, B, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2004-06-03 | | Release date: | 2005-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Lumazine Synthase from Mycobacterium Tuberculosis as a Target for Rational Drug Design: Binding Mode of a New Class of Purinetrione Inhibitors(,)
Biochemistry, 44, 2005
|
|
5ICV
 
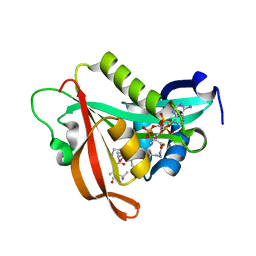 | | Crystal structure of human NatF (hNaa60) bound to a bisubstrate analogue | | Descriptor: | MET-LYS-ALA-VAL-LIG, N-alpha-acetyltransferase 60, [5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)furan-2-yl]methyl (3R)-4-{[3-({(E)-2-[(2,2-dihydroxyethyl)sulfanyl]ethenyl}amino)-3-oxopropyl]amino}-3-hydroxy-2,2-dimethyl-4-oxobutyl dihydrogen diphosphate | | Authors: | Stove, S.I, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal Structure of the Golgi-Associated Human N alpha-Acetyltransferase 60 Reveals the Molecular Determinants for Substrate-Specific Acetylation.
Structure, 24, 2016
|
|
1W29
 
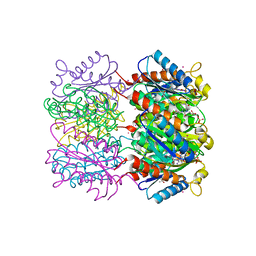 | | Lumazine Synthase from Mycobacterium tuberculosis bound to 3-(1,3,7- trihydro-9-D-ribityl-2,6,8-purinetrione-7-yl)butane 1-phosphate | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 4-{2,6,8-TRIOXO-9-[(2R,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL]-1,2,3,6,8,9-HEXAHYDRO-7H-PURIN-7-YL}BUTYL DIHYDROGEN PHOSPHATE, 4-{2,6,8-TRIOXO-9-[(2S,3R,4R)-2,3,4,5-TETRAHYDROXYPENTYL]-1,2,3,6,8,9-HEXAHYDRO-7H-PURIN-7-YL}BUTYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Morgunova, E, Meining, W, Illarionov, B, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2004-07-01 | | Release date: | 2005-03-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Lumazine Synthase from Mycobacterium Tuberculosis as a Target for Rational Drug Design: Binding Mode of a New Class of Purinetrione Inhibitors(,)
Biochemistry, 44, 2005
|
|
8RWJ
 
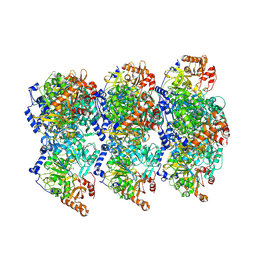 | | cryoEM structure of Acs1 filament determined by FilamentID | | Descriptor: | Acetyl-coenzyme A synthetase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] ethanoate | | Authors: | Hugener, J, Xu, J, Wettstein, R, Ioannidi, L, Velikov, D, Wollweber, F, Henggeler, A, Matos, J, Pilhofer, M. | | Deposit date: | 2024-02-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | FilamentID reveals the composition and function of metabolic enzyme polymers during gametogenesis.
Cell, 187, 2024
|
|
4U9W
 
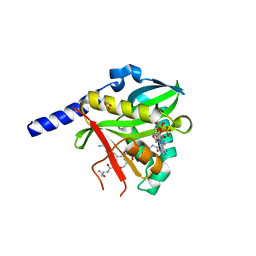 | | Crystal Structure of NatD bound to H4/H2A peptide and CoA | | Descriptor: | COENZYME A, GLYCEROL, Histone H4/H2A N-terminus, ... | | Authors: | Magin, R.S, Liszczak, G.P, Marmorstein, R. | | Deposit date: | 2014-08-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The Molecular Basis for Histone H4- and H2A-Specific Amino-Terminal Acetylation by NatD.
Structure, 23, 2015
|
|
8RWK
 
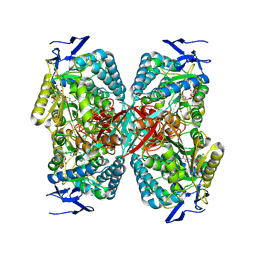 | | cryoEM structure of the central Ald4 filament determined by FilamentID | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium-activated aldehyde dehydrogenase, mitochondrial | | Authors: | Hugener, J, Xu, J, Wettstein, R, Ioannidi, L, Velikov, D, Wollweber, F, Henggeler, A, Matos, J, Pilhofer, M. | | Deposit date: | 2024-02-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | FilamentID reveals the composition and function of metabolic enzyme polymers during gametogenesis.
Cell, 187, 2024
|
|
4U9V
 
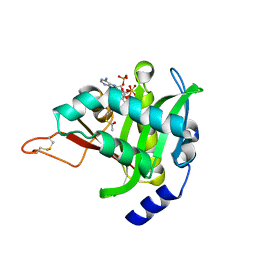 | |
4UA3
 
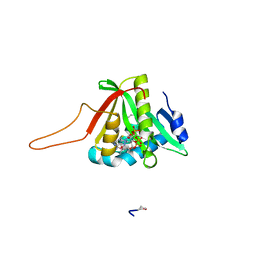 | |
1GMY
 
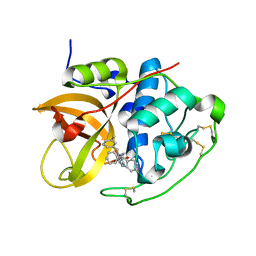 | | Cathepsin B complexed with dipeptidyl nitrile inhibitor | | Descriptor: | 2-AMINOETHANIMIDIC ACID, 3-METHYLPHENYLALANINE, CATHEPSIN B, ... | | Authors: | Greenspan, P.D, Clark, K.L, Tommasi, R.A, Cowen, S.D, McQuire, L.W, Farley, D.L, van Duzer, J.H, Goldberg, R.L, Zhou, H, Du, Z, Fitt, J.J, Coppa, D.E, Fang, Z, Macchia, W, Zhu, L, Capparelli, M.P, Goldstein, R, Wigg, A.M, Doughty, J.R, Bohacek, R.S, Knap, A.K. | | Deposit date: | 2001-09-25 | | Release date: | 2002-09-19 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of Dipeptidyl Nitriles as Potent and Selective Inhibitors of Cathepsin B Through Structure-Based Drug Design
J.Med.Chem., 44, 2001
|
|
5ICW
 
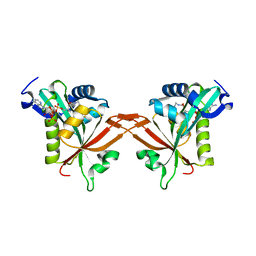 | | Crystal structure of human NatF (hNaa60) homodimer bound to Coenzyme A | | Descriptor: | CHLORIDE ION, COENZYME A, N-alpha-acetyltransferase 60 | | Authors: | Stove, S.I, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystal Structure of the Golgi-Associated Human N alpha-Acetyltransferase 60 Reveals the Molecular Determinants for Substrate-Specific Acetylation.
Structure, 24, 2016
|
|
1IHB
 
 | | CRYSTAL STRUCTURE OF P18-INK4C(INK6) | | Descriptor: | CYCLIN-DEPENDENT KINASE 6 INHIBITOR | | Authors: | Ravichandran, V, Swaminathan, K, Marmorstein, R. | | Deposit date: | 1997-10-25 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the CDK4/6 inhibitory protein p18INK4c provides insights into ankyrin-like repeat structure/function and tumor-derived p16INK4 mutations.
Nat.Struct.Biol., 5, 1998
|
|
1YK4
 
 | | Ultra-high resolution structure of Pyrococcus abyssi rubredoxin W4L/R5S | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Bonisch, H, Schmidt, C.L, Bianco, P, Ladenstein, R. | | Deposit date: | 2005-01-17 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (0.69 Å) | | Cite: | Ultrahigh-resolution study on Pyrococcus abyssi rubredoxin. I. 0.69 A X-ray structure of mutant W4L/R5S.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YK5
 
 | | Pyrococcus abyssi rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Bonisch, H, Schmidt, C.L, Bianco, P, Ladenstein, R. | | Deposit date: | 2005-01-17 | | Release date: | 2006-01-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Ultrahigh-resolution study on Pyrococcus abyssi rubredoxin. I. 0.69 A X-ray structure of mutant W4L/R5S.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1RVV
 
 | | SYNTHASE/RIBOFLAVIN SYNTHASE COMPLEX OF BACILLUS SUBTILIS | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, PHOSPHATE ION, RIBOFLAVIN SYNTHASE | | Authors: | Ritsert, K, Huber, R, Turk, D, Ladenstein, R, Schmidt-Baese, K, Bacher, A. | | Deposit date: | 1995-10-25 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Studies on the lumazine synthase/riboflavin synthase complex of Bacillus subtilis: crystal structure analysis of reconstituted, icosahedral beta-subunit capsids with bound substrate analogue inhibitor at 2.4 A resolution.
J.Mol.Biol., 253, 1995
|
|
5DFP
 
 | | Crystal structure of PAK1 in complex with an inhibitor compound FRAX1036 | | Descriptor: | 6-[2-chloro-4-(6-methylpyrazin-2-yl)phenyl]-8-ethyl-2-{[2-(1-methylpiperidin-4-yl)ethyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, DIMETHYL SULFOXIDE, Serine/threonine-protein kinase PAK 1 | | Authors: | Maksimoska, J, Marmorstein, R, Wang, W. | | Deposit date: | 2015-08-27 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of Selective PAK1 Inhibitor G-5555: Improving Properties by Employing an Unorthodox Low-pK a Polar Moiety.
Acs Med.Chem.Lett., 6, 2015
|
|
2RC4
 
 | | Crystal Structure of the HAT domain of the human MOZ protein | | Descriptor: | ACETYL COENZYME *A, Histone acetyltransferase MYST3, ZINC ION | | Authors: | Holbert, M.A, Sikorski, T, Snowflack, D, Marmorstein, R. | | Deposit date: | 2007-09-19 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The human monocytic leukemia zinc finger histone acetyltransferase domain contains DNA-binding activity implicated in chromatin targeting.
J.Biol.Chem., 282, 2007
|
|
2C4E
 
 | | Crystal Structure of Methanocaldococcus jannaschii Nucleoside Kinase - An Archaeal Member of the Ribokinase Family | | Descriptor: | MAGNESIUM ION, SUGAR KINASE MJ0406 | | Authors: | Arnfors, L, Hansen, T, Meining, W, Schoenheit, P, Ladenstein, R. | | Deposit date: | 2005-10-18 | | Release date: | 2006-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Methanocaldococcus Jannaschii Nucleoside Kinase: An Archaeal Member of the Ribokinase Family.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2C49
 
 | | Crystal Structure of Methanocaldococcus jannaschii Nucleoside Kinase - An Archaeal Member of the Ribokinase Family | | Descriptor: | ADENOSINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Arnfors, L, Hansen, T, Meining, W, Schoenheit, P, Ladenstein, R. | | Deposit date: | 2005-10-17 | | Release date: | 2006-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of Methanocaldococcus Jannaschii Nucleoside Kinase: An Archaeal Member of the Ribokinase Family.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2TMG
 
 | | THERMOTOGA MARITIMA GLUTAMATE DEHYDROGENASE MUTANT S128R, T158E, N117R, S160E | | Descriptor: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | Authors: | Knapp, S, Lebbink, J.H.G, van der Oost, J, de Vos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering activity and stability of Thermotoga maritima glutamate dehydrogenase. II: construction of a 16-residue ion-pair network at the subunit interface.
J.Mol.Biol., 289, 1999
|
|
2X0I
 
 | | 2.9 A RESOLUTION STRUCTURE OF MALATE DEHYDROGENASE FROM ARCHAEOGLOBUS FULGIDUS IN COMPLEX WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MALATE DEHYDROGENASE, SODIUM ION, ... | | Authors: | Irimia, A, Madern, D, Zaccai, G, Vellieux, F.M.D, Karshikoff, A, Tibbelin, G, Ladenstein, R, Lien, T, Birkeland, N.-K. | | Deposit date: | 2009-12-14 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | The 2.9A Resolution Crystal Structure of Malate Dehydrogenase from Archaeoglobus Fulgidus: Mechanisms of Oligomerisation and Thermal Stabilisation.
J.Mol.Biol., 335, 2004
|
|
2R7G
 
 | |
2C9D
 
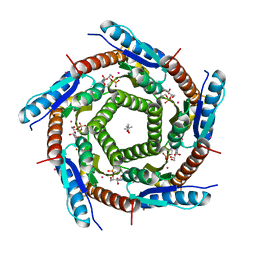 | | Lumazine Synthase from Mycobacterium tuberculosis Bound to 3-(1,3,7- TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL)HEXANE 1-PHOSPHATE | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(1,3,7-TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL ) HEXANE 1-PHOSPHATE, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, ... | | Authors: | Morgunova, E, Illarionov, B, Jin, G, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Thermodynamic Insights Into the Binding Mode of Five Novel Inhibitors of Lumazine Synthase from Mycobacterium Tuberculosis.
FEBS J., 273, 2006
|
|
