6C6R
 
 | |
6C6P
 
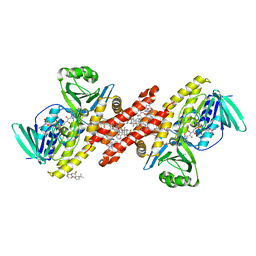 | | Human squalene epoxidase (SQLE, squalene monooxygenase) structure with FAD and NB-598 | | Descriptor: | (2E)-N-({3-[([3,3'-bithiophen]-5-yl)methoxy]phenyl}methyl)-N-ethyl-6,6-dimethylhept-2-en-4-yn-1-amine, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Padyana, A.K, Jin, L. | | Deposit date: | 2018-01-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and inhibition mechanism of the catalytic domain of human squalene epoxidase.
Nat Commun, 10, 2019
|
|
7BNR
 
 | |
7BNK
 
 | |
3KAF
 
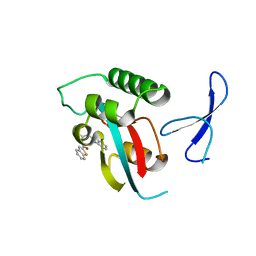 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 3-(1H-benzimidazol-2-yl)-N-(1-benzothiophen-2-ylcarbonyl)-D-alanine, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KAI
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | (2R)-2-[(2-methyl-5-phenyl-pyrazol-3-yl)carbonylamino]-3-naphthalen-2-yl-propanoic acid, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KAH
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 3-(1H-benzimidazol-2-yl)-N-[(1-methyl-3-phenyl-1H-pyrazol-5-yl)carbonyl]-D-alanine, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1REC
 
 | | THREE-DIMENSIONAL STRUCTURE OF RECOVERIN, A CALCIUM SENSOR IN VISION | | Descriptor: | CALCIUM ION, RECOVERIN | | Authors: | Flaherty, K.M, Zozulya, S, Stryer, L, Mckay, D.B. | | Deposit date: | 1993-10-29 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Structure of Recoverin, a Calcium Sensor in Vision
Cell(Cambridge,Mass.), 75, 1993
|
|
3KAD
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 3-(1H-benzimidazol-2-yl)-N-(3-phenylpropanoyl)-D-alanine, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KAB
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 6-methyl-1H-indole-2-carboxylic acid, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KAC
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 3-(1H-benzimidazol-2-yl)propanoic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1DRK
 
 | |
1DRJ
 
 | |
1E57
 
 | |
1DBP
 
 | |
7R26
 
 | | PI3K delta in complex with SD5 | | Descriptor: | 5-[2,6-di(morpholin-4-yl)pyrimidin-4-yl]-4-(trifluoromethyl)pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Gutmann, S, Rummel, G, Shrestha, B. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor.
J.Med.Chem., 65, 2022
|
|
7R2B
 
 | | PI3Kdelta in complex with an inhibitor | | Descriptor: | (4~{S})-3-[6-[2-azanyl-4-(trifluoromethyl)pyrimidin-5-yl]-2-morpholin-4-yl-pyrimidin-4-yl]-4-methyl-1,3-oxazolidin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Gutmann, S, Rummel, G, Shrestha, B. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor.
J.Med.Chem., 65, 2022
|
|
6N0M
 
 | |
4C8B
 
 | | Structure of the kinase domain of human RIPK2 in complex with ponatinib | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Canning, P, Krojer, T, Bradley, A, Mahajan, P, Goubin, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Inflammatory Signaling by NOD-RIPK2 Is Inhibited by Clinically Relevant Type II Kinase Inhibitors.
Chem. Biol., 22, 2015
|
|
4S0H
 
 | | TBX5 DB, NKX2.5 HD, ANF DNA Complex | | Descriptor: | 5'-D(*CP*CP*AP*CP*TP*TP*CP*AP*AP*AP*GP*GP*TP*GP*TP*GP*AP*GP*A)-3', 5'-D(*TP*CP*TP*CP*AP*CP*AP*CP*CP*TP*TP*TP*GP*AP*AP*GP*TP*GP*G)-3', Homeobox protein Nkx-2.5, ... | | Authors: | Pradhan, L. | | Deposit date: | 2014-12-31 | | Release date: | 2015-12-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | Intermolecular Interactions of Cardiac Transcription Factors NKX2.5 and TBX5.
Biochemistry, 55, 2016
|
|
8CA0
 
 | |
7TZ7
 
 | | PI3K alpha in complex with an inhibitor | | Descriptor: | (4S,5R)-3-[2'-amino-2-(morpholin-4-yl)-4'-(trifluoromethyl)[4,5'-bipyrimidin]-6-yl]-4-(hydroxymethyl)-5-methyl-1,3-oxazolidin-2-one, Isoform 3 of Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Tang, J. | | Deposit date: | 2022-02-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor.
J.Med.Chem., 65, 2022
|
|
8ORF
 
 | |
8ORE
 
 | |
8ORG
 
 | |
