4O63
 
 | | Co-enzyme Induced Conformational Changes in Bovine Eye Glyceraldehyde 3-Phosphate Dehydrogenase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Baker, B.Y, Shi, W, Wang, B, Palczewski, K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules.
Protein Sci., 23, 2014
|
|
4NN0
 
 | | Crystal structure of the C1QTNF5 globular domain in space group P63 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Complement C1q tumor necrosis factor-related protein 5, ... | | Authors: | Tu, X, Palczewski, K. | | Deposit date: | 2013-11-15 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The macular degeneration-linked C1QTNF5 (S163) mutation causes higher-order structural rearrangements.
J.Struct.Biol., 186, 2014
|
|
3FSN
 
 | | Crystal structure of RPE65 at 2.14 angstrom resolution | | Descriptor: | FE (II) ION, Retinal pigment epithelium-specific 65 kDa protein, TETRAETHYLENE GLYCOL | | Authors: | Kiser, P.D, Lodowski, D.T, Palczewski, K. | | Deposit date: | 2009-01-11 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of native RPE65, the retinoid isomerase of the visual cycle.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2I37
 
 | | Crystal structure of a photoactivated rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Rhodopsin, ... | | Authors: | Lodowski, D.T, Stenkamp, R.E, Salom, D, Le Trong, I, Palczewski, K. | | Deposit date: | 2006-08-17 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Crystal structure of a photoactivated deprotonated intermediate of rhodopsin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I36
 
 | | Crystal structure of trigonal crystal form of ground-state rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, Rhodopsin, ... | | Authors: | Stenkamp, R.E, Le Trong, I, Lodowski, D.T, Salom, D, Palczewski, K. | | Deposit date: | 2006-08-17 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of a photoactivated deprotonated intermediate of rhodopsin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1GNW
 
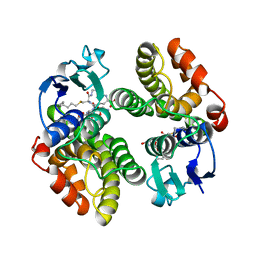 | | STRUCTURE OF GLUTATHIONE S-TRANSFERASE | | Descriptor: | GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Reinemer, P, Prade, L, Hof, P, Neuefeind, T, Huber, R, Palme, K, Bartunik, H.D, Bieseler, B. | | Deposit date: | 1996-09-15 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of glutathione S-transferase from Arabidopsis thaliana at 2.2 A resolution: structural characterization of herbicide-conjugating plant glutathione S-transferases and a novel active site architecture.
J.Mol.Biol., 255, 1996
|
|
3VAT
 
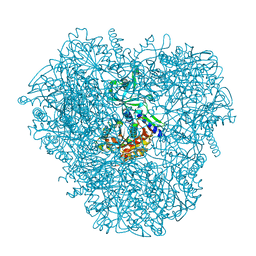 | | Crystal structure of DNPEP, ZnMg form | | Descriptor: | Aspartyl aminopeptidase, MAGNESIUM ION, ZINC ION | | Authors: | Kiser, P.D, Chen, Y, Palczewski, K. | | Deposit date: | 2011-12-29 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into substrate specificity and metal activation of Mammalian tetrahedral aspartyl aminopeptidase.
J.Biol.Chem., 287, 2012
|
|
3VAR
 
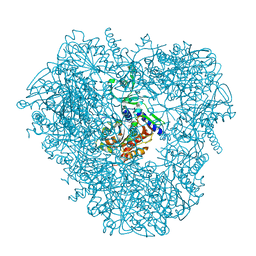 | | Crystal structure of DNPEP, ZnZn form | | Descriptor: | Aspartyl aminopeptidase, ZINC ION | | Authors: | Kiser, P.D, Chen, Y, Palczewski, K. | | Deposit date: | 2011-12-29 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights into substrate specificity and metal activation of Mammalian tetrahedral aspartyl aminopeptidase.
J.Biol.Chem., 287, 2012
|
|
1HZX
 
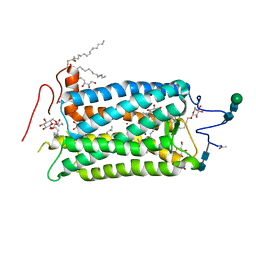 | | CRYSTAL STRUCTURE OF BOVINE RHODOPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Teller, D.C, Okada, T, Behnke, C.A, Palczewski, K, Stenkamp, R.E. | | Deposit date: | 2001-01-26 | | Release date: | 2001-07-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs).
Biochemistry, 40, 2001
|
|
2I35
 
 | | Crystal structure of rhombohedral crystal form of ground-state rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, RETINAL, ... | | Authors: | Stenkamp, R.E, Le Trong, I, Lodowski, D.T, Salom, D, Palczewski, K. | | Deposit date: | 2006-08-17 | | Release date: | 2006-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of a photoactivated deprotonated intermediate of rhodopsin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4F3A
 
 | |
4F3J
 
 | |
4F30
 
 | |
4F3D
 
 | |
4F2Z
 
 | |
4DPZ
 
 | | Crystal structure of human HRASLS2 | | Descriptor: | HRAS-like suppressor 2 | | Authors: | Kiser, P.D, Golczak, M, Sears, A.E, Lodowski, D.T, Palczewski, K. | | Deposit date: | 2012-02-14 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis for the Acyltransferase Activity of Lecithin:Retinol Acyltransferase-like Proteins.
J.Biol.Chem., 287, 2012
|
|
4DOT
 
 | | Crystal structure of human HRASLS3. | | Descriptor: | Group XVI phospholipase A2 | | Authors: | Kiser, P.D, Golczak, M, Sears, A.E, Lodowski, D.T, Palczewski, K. | | Deposit date: | 2012-02-10 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for the Acyltransferase Activity of Lecithin:Retinol Acyltransferase-like Proteins.
J.Biol.Chem., 287, 2012
|
|
3BZG
 
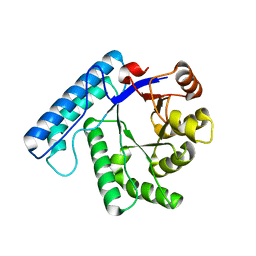 | | UVDE pH4.4 | | Descriptor: | UV endonuclease | | Authors: | Meulenbroek, E.M, Paspaleva, K, Thomassen, E.A.J, Abrahams, J.P, Goosen, N, Pannu, N.S. | | Deposit date: | 2008-01-18 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Involvement of a carboxylated lysine in UV damage endonuclease
Protein Sci., 18, 2009
|
|
2NWC
 
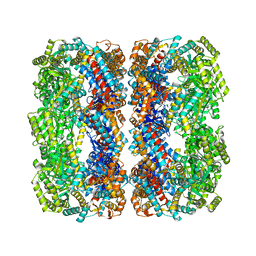 | |
6B20
 
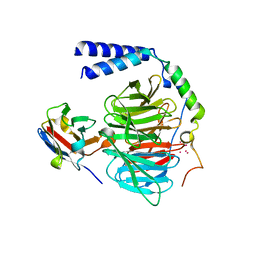 | | Crystal structure of a complex between G protein beta gamma dimer and an inhibitory Nanobody regulator | | Descriptor: | CHLORIDE ION, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, ... | | Authors: | Gulati, S, Kiser, P.D, Palczewski, K. | | Deposit date: | 2017-09-19 | | Release date: | 2018-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Targeting G protein-coupled receptor signaling at the G protein level with a selective nanobody inhibitor.
Nat Commun, 9, 2018
|
|
5QTZ
 
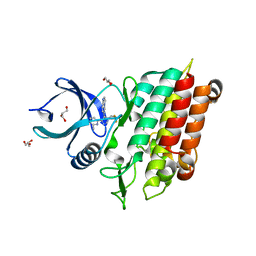 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) IN COMPLEX WITH 6-[1-(2,2-DIFLUOROETHYL)-4-(6-METHYLPYRIDIN-2-YL)-1H-IMIDAZOL-5-YL]IMIDAZO[1,2-A]PYRIDINE | | Descriptor: | 6-[1-(2,2-difluoroethyl)-4-(6-methylpyridin-2-yl)-1H-imidazol-5-yl]imidazo[1,2-a]pyridine, GLYCEROL, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of BMS-986260, a Potent, Selective, and Orally Bioavailable TGF beta R1 Inhibitor as an Immuno-oncology Agent.
Acs Med.Chem.Lett., 11, 2020
|
|
5QU0
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) IN COMPLEX WITH 6-[4-(3-CHLORO-4-FLUOROPHENYL)-1-(2-HYDROXYETHYL)-1H-IMIDAZOL-5-YL]IMIDAZO[1,2-B]PYRIDAZINE-3-CARBONITRILE | | Descriptor: | 6-[4-(3-chloro-4-fluorophenyl)-1-(2-hydroxyethyl)-1H-imidazol-5-yl]imidazo[1,2-b]pyridazine-3-carbonitrile, GLYCEROL, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of BMS-986260, a Potent, Selective, and Orally Bioavailable TGF beta R1 Inhibitor as an Immuno-oncology Agent.
Acs Med.Chem.Lett., 11, 2020
|
|
6VQL
 
 | |
1EKR
 
 | | MOAC PROTEIN FROM E. COLI | | Descriptor: | MOLYBDENUM COFACTOR BIOSYNTHESIS PROTEIN C | | Authors: | Schindelin, H, Liu, M.T.W, Wuebbens, M.M, Rajagopalan, K.V. | | Deposit date: | 2000-03-09 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into molybdenum cofactor deficiency provided by the crystal structure of the molybdenum cofactor biosynthesis protein MoaC.
Structure Fold.Des., 8, 2000
|
|
1EKS
 
 | | ASP128ALA VARIANT OF MOAC PROTEIN FROM E. COLI | | Descriptor: | L(+)-TARTARIC ACID, MOLYBDENUM COFACTOR BIOSYNTHESIS PROTEIN C | | Authors: | Schindelin, H, Liu, M.T.W, Wuebbens, M.M, Rajagopalan, K.V. | | Deposit date: | 2000-03-09 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into molybdenum cofactor deficiency provided by the crystal structure of the molybdenum cofactor biosynthesis protein MoaC.
Structure Fold.Des., 8, 2000
|
|
