1YJ2
 
 | | Cyclized, non-dehydrated post-translational product for S65A Y66S H148G GFP variant | | Descriptor: | 1,2-ETHANEDIOL, Green Fluorescent Protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-01-13 | | Release date: | 2005-02-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Understanding GFP Chromophore Biosynthesis: Controlling Backbone Cyclization and Modifying Post-translational Chemistry.
Biochemistry, 44, 2005
|
|
1YJF
 
 | | Cyclized post-translational product for S65A Y66S (GFPhal) green fluorescent protein variant | | Descriptor: | Green Fluorescent Protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Understanding GFP Chromophore Biosynthesis: Controlling Backbone Cyclization and Modifying Post-translational Chemistry.
Biochemistry, 44, 2005
|
|
7KG3
 
 | | Crystal structure of CoV-2 Nsp3 Macrodomain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MALONATE ION, ... | | Authors: | Arvai, A, Brosey, C.A, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-15 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
3RD2
 
 | | NIP45 SUMO-like Domain 2 | | Descriptor: | NFATC2-interacting protein | | Authors: | Perry, J.J.P, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA repair and global sumoylation are regulated by distinct Ubc9 noncovalent complexes.
Mol.Cell.Biol., 31, 2011
|
|
7KXB
 
 | | Crystal structure of SARS-CoV-2 Nsp3 Macrodomain complex with PARG329 | | Descriptor: | BETA-MERCAPTOETHANOL, N-{3-[(1,3-dimethyl-2,6-dioxo-2,3,6,9-tetrahydro-1H-purin-8-yl)sulfanyl]propyl}-N'-[2-(morpholin-4-yl)ethyl]thiourea, Non-structural protein 3, ... | | Authors: | Arvai, A, Brosey, C.A, Bommagani, S, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-12-03 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KG1
 
 | | Structure of human PARG complexed with PARG-002 | | Descriptor: | 1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]amino}-3,7-dihydro-1H-purine-2,6-dione, CACODYLATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Brosey, C.A, Balapiti-Modarage, L.P.F, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-15 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KG7
 
 | | Structure of human PARG complexed with PARG-292 | | Descriptor: | 8-{[2-(1,1-dioxo-1lambda~6~,4-thiazinan-4-yl)ethyl]sulfanyl}-1,3-dimethyl-3,7-dihydro-1H-purine-2,6-dione, DIMETHYL SULFOXIDE, Poly(ADP-ribose) glycohydrolase, ... | | Authors: | Brosey, C.A, Balapiti-Modarage, L.P.F, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KFP
 
 | | Structure of human PARG complexed with PARG-119 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{[2-(1,3-dimethyl-2-oxo-6-sulfanylidene-1,2,3,6-tetrahydro-7H-purin-7-yl)ethyl]carbamoyl}methanesulfonamide, ... | | Authors: | Brosey, C.A, Bommagani, S, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KG0
 
 | | Structure of human PARG complexed with PARG-131 | | Descriptor: | 1,2-ETHANEDIOL, 5-({4-[(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)methyl]phenyl}methyl)pyrimidine-2,4,6(1H,3H,5H)-trione, Poly(ADP-ribose) glycohydrolase, ... | | Authors: | Arvai, A, Bommagani, S, Brosey, C.A, Jones, D.E, Warden, L.S, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-15 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KG6
 
 | | Structure of human PARG complexed with PARG-322 | | Descriptor: | 1-{2-[(1,3-dimethyl-2,6-dioxo-2,3,6,7-tetrahydro-1H-purin-8-yl)sulfanyl]ethyl}piperidine-4-carboxylic acid, Poly(ADP-ribose) glycohydrolase | | Authors: | Brosey, C.A, Balapiti-Modarage, L.P.F, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KG8
 
 | | Structure of human PARG complexed with PARG-061 | | Descriptor: | 1,3-dimethyl-8-{[2-(morpholin-4-yl)-2-oxoethyl]sulfanyl}-6-sulfanylidene-1,3,6,7-tetrahydro-2H-purin-2-one, CACODYLATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Brosey, C.A, Balapiti-Modarage, L.P.F, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
1YAI
 
 | | X-RAY STRUCTURE OF A BACTERIAL COPPER,ZINC SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER, ZINC SUPEROXIDE DISMUTASE, ... | | Authors: | Bourne, Y, Redford, S.M, Lo, T.P, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 1996-02-03 | | Release date: | 1997-08-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel dimeric interface and electrostatic recognition in bacterial Cu,Zn superoxide dismutase.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1YHG
 
 | |
7LG7
 
 | | Crystal structure of CoV-2 Nsp3 Macrodomain complex with PARG345 | | Descriptor: | 3-[(1,3-dimethyl-2,6-dioxo-2,3,6,9-tetrahydro-1H-purin-8-yl)sulfanyl]-N-{[2-(morpholin-4-yl)ethyl]sulfonyl}propanamide, Non-structural protein 3, SULFATE ION | | Authors: | Arvai, A, Brosey, C.A, Bommagani, S, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2021-01-19 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
1YHH
 
 | |
1DWW
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and dihydrobiopterin | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
1DWX
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and tetrahydrobiopterin | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
1DWV
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and 4-amino tetrahydrobiopterin | | Descriptor: | 2,4-DIAMINO-6-[2,3-DIHYDROXY-PROP-3-YL]-5,6,7,8-TETRAHYDROPTERIDINE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
3F7K
 
 | | X-ray Crystal Structure of an Alvinella pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Copper,Zinc Superoxide Dismutase, ... | | Authors: | Shin, D.S, DiDonato, M, Barondeau, D.P, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2008-11-09 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Superoxide Dismutase from the Eukaryotic Thermophile Alvinella pompejana: Structures, Stability, Mechanism, and Insights into Amyotrophic Lateral Sclerosis.
J.Mol.Biol., 385, 2009
|
|
3F7L
 
 | | X-ray Crystal Structure of Alvinella pompejana Cu,Zn Superoxide Dismutase | | Descriptor: | ACETIC ACID, COPPER (I) ION, COPPER (II) ION, ... | | Authors: | Shin, D.S, DiDonato, M, Barondeau, D.P, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2008-11-09 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Superoxide Dismutase from the Eukaryotic Thermophile Alvinella pompejana: Structures, Stability, Mechanism, and Insights into Amyotrophic Lateral Sclerosis.
J.Mol.Biol., 385, 2009
|
|
7LW7
 
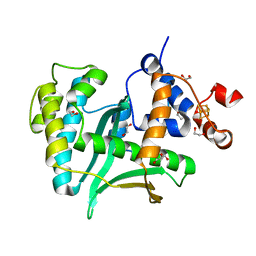 | | Human Exonuclease 5 crystal structure | | Descriptor: | 1,2-ETHANEDIOL, Exonuclease V, GLYCEROL, ... | | Authors: | Tsai, C.L, Tainer, J.A. | | Deposit date: | 2021-02-27 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | EXO5-DNA structure and BLM interactions direct DNA resection critical for ATR-dependent replication restart.
Mol.Cell, 81, 2021
|
|
7LW9
 
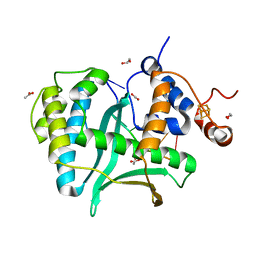 | | Human Exonuclease 5 crystal structure in complex with ssDNA, Sm, and Na | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*AP*TP*TP*GP*CP*TP*GP*AP*AP*GP*GP*G)-3'), ... | | Authors: | Tsai, C.L, Tainer, J.A. | | Deposit date: | 2021-02-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | EXO5-DNA structure and BLM interactions direct DNA resection critical for ATR-dependent replication restart.
Mol.Cell, 81, 2021
|
|
7LW8
 
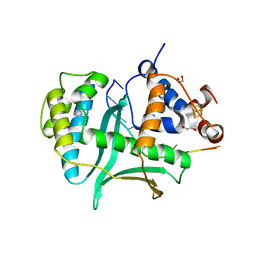 | | Human Exonuclease 5 crystal structure in complex with a ssDNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Exonuclease V, ... | | Authors: | Tsai, C.L, Tainer, J.A. | | Deposit date: | 2021-02-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | EXO5-DNA structure and BLM interactions direct DNA resection critical for ATR-dependent replication restart.
Mol.Cell, 81, 2021
|
|
7LPZ
 
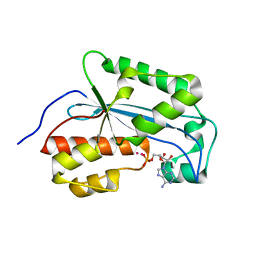 | |
7LQ0
 
 | |
