5N2C
 
 | | Crystal structure of the peptidoglycan-associated lipoprotein from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ACETATE ION, ... | | Authors: | Matterazzo, E, Bolognesi, M, Gourlay, L.J. | | Deposit date: | 2017-02-07 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designing Probes for Immunodiagnostics: Structural Insights into an Epitope Targeting Burkholderia Infections.
ACS Infect Dis, 3, 2017
|
|
1SWM
 
 | | X-RAY CRYSTAL STRUCTURE OF THE FERRIC SPERM WHALE MYOGLOBIN: IMIDAZOLE COMPLEX AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | AZIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rizzi, M, Ascenzi, P, Coda, A, Brunori, M, Bolognesi, M. | | Deposit date: | 1992-03-03 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of the ferric sperm whale myoglobin: imidazole complex at 2.0 A resolution.
J.Mol.Biol., 217, 1991
|
|
4O4R
 
 | | Murine Norovirus RdRp in complex with PPNDS | | Descriptor: | 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, RNA-dependent-RNA-polymerase, SULFATE ION | | Authors: | Croci, R, Tarantino, D, Milani, M, Pezzullo, M, Bolognesi, M, Mastrangelo, E. | | Deposit date: | 2013-12-19 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PPNDS inhibits murine Norovirus RNA-dependent RNA-polymerase mimicking two RNA stacking bases.
Febs Lett., 588, 2014
|
|
7ZPA
 
 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZLA
 
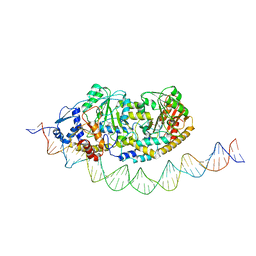 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Savino, C, Exertier, C, Bolognesi, M, Chaves Sanjuan, A. | | Deposit date: | 2022-04-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZN5
 
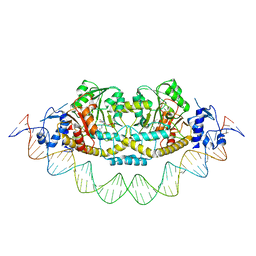 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry. | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZTH
 
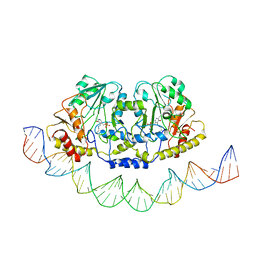 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-05-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
1OAJ
 
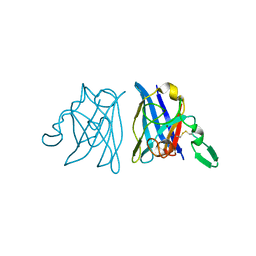 | | Active site copper and zinc ions modulate the quaternary structure of prokaryotic Cu,Zn superoxide dismutase | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Cioni, P, Pesce, A, Rocca, B.M.D, Castelli, S, Falconi, M, Parrilli, L, Bolognesi, M, Strambini, G, Desideri, A. | | Deposit date: | 2003-01-14 | | Release date: | 2003-02-27 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Active-Site Copper and Zinc Ions Modulate the Quaternary Structure of Prokaryotic Cu,Zn Superoxide Dismutase
J.Mol.Biol., 326, 2003
|
|
4NRU
 
 | | Murine Norovirus RNA-dependent-RNA-polymerase in complex with Compound 6, a suramin derivative | | Descriptor: | 4-({4-methyl-3-[(3-nitrobenzoyl)amino]benzoyl}amino)naphthalene-1,5-disulfonic acid, MAGNESIUM ION, RNA dependent RNA polymerase | | Authors: | Milani, M, Croci, R, Pezzullo, M, Tarantino, D, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2013-11-27 | | Release date: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural bases of norovirus RNA dependent RNA polymerase inhibition by novel suramin-related compounds.
Plos One, 9, 2014
|
|
2VG6
 
 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-bromophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
2VG5
 
 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-chlorophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
1OJ6
 
 | | Human brain neuroglobin three-dimensional structure | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Pesce, A, Dewilde, S, Nardini, M, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Bolognesi, M. | | Deposit date: | 2003-07-03 | | Release date: | 2003-09-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human Brain Neuroglobin Structure Reveals a Distinct Mode of Controlling Oxygen Affinity
Structure, 11, 2003
|
|
4B3W
 
 | | Crystal structure of human cytoglobin H(E7)Q mutant | | Descriptor: | ACETATE ION, CYANIDE ION, CYTOGLOBIN, ... | | Authors: | Gabba, M, Abbruzzetti, S, Spyrakis, F, Forti, F, Bruno, S, Mozzarelli, A, Luque, F.J, Viappiani, C, Cozzini, P, Nardini, M, Germani, F, Bolognesi, M, Moens, L, Dewilde, S. | | Deposit date: | 2012-07-26 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Co Rebinding Kinetics and Molecular Dynamics Simulations Highlight Dynamic Regulation of Internal Cavities in Human Cytoglobin.
Plos One, 8, 2013
|
|
2VG7
 
 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-iodophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
5N2B
 
 | |
1OAL
 
 | | Active site copper and zinc ions modulate the quaternary structure of prokaryotic Cu,Zn superoxide dismutase | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Cioni, P, Pesce, A, Rocca, B.M.D, Castellifalconiparrilli, L, Bolognesi, M, Strambini, G, Desideri, A. | | Deposit date: | 2003-01-15 | | Release date: | 2003-02-27 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active-Site Copper and Zinc Ions Modulate the Quaternary Structure of Prokaryotic Cu,Zn Superoxide Dismutase
J.Mol.Biol., 326, 2003
|
|
1OLL
 
 | | Extracellular region of the human receptor NKp46 | | Descriptor: | 1,2-ETHANEDIOL, NK RECEPTOR | | Authors: | Ponassi, M, Cantoni, C, Biassoni, R, Conte, R, Spallarossa, A, Pesce, A, Moretta, A, Moretta, L, Bolognesi, M, Bordo, D. | | Deposit date: | 2003-08-07 | | Release date: | 2003-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of the Human Nk Cell Triggering Receptor Nkp46 Ectodomain
Biochem.Biophys.Res.Commun., 309, 2003
|
|
1UMA
 
 | | ALPHA-THROMBIN (HIRUGEN) COMPLEXED WITH NA-(N,N-DIMETHYLCARBAMOYL)-ALPHA-AZALYSINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN, HIRUDIN I, ... | | Authors: | Nardini, M, Pesce, A, Rizzi, M, Casale, E, Ferraccioli, R, Balliano, G, Milla, P, Ascenzi, P, Bolognesi, M. | | Deposit date: | 1996-03-26 | | Release date: | 1996-11-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the active site titrant N alpha-(N,N-dimethylcarbamoyl)-alpha-azalysine p-nitrophenyl ester: a comparative kinetic and X-ray crystallographic study.
J.Mol.Biol., 258, 1996
|
|
4CFI
 
 | | 3D structure of FliC from Burkholderia pseudomallei | | Descriptor: | FLAGELLIN | | Authors: | Lassaux, P, Peri, C, Ferrer-Navarro, M, Gourlay, L.J, Conchillo-Sole, O, Daura, X, Colombo, G, Bolognesi, M. | | Deposit date: | 2013-11-18 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Sequence- and Structure-Based Immunoreactive Epitope Discovery for Burkholderia Pseudomallei Flagellin.
Plos Negl Trop Dis, 9, 2015
|
|
2VYW
 
 | | Hemoglobin (Hb2) from trematode Fasciola hepatica | | Descriptor: | HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dewilde, S, Ioanitescu, A.I, Kiger, L, Gilany, K, Marden, M.C, Van Doorslaer, S, Vercruysse, J, Pesce, A, Nardini, M, Bolognesi, M, Moens, L. | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Hemoglobins of the Trematodes Fasciola Hepatica and Paramphistomum Epiclitum: A Molecular Biological, Physico-Chemical, Kinetic, and Vaccination Study.
Protein Sci., 17, 2008
|
|
2XTL
 
 | | Structure of the major pilus backbone protein from Streptococcus Agalactiae | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, POTASSIUM ION | | Authors: | Rinauda, D, Gourlay, L.J, Soriano, M, Grandi, G, Bolognesi, M. | | Deposit date: | 2010-10-11 | | Release date: | 2011-07-06 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Approach to Rationally Design a Chimeric Protein for an Effective Vaccine Against Group B Streptococcus Infections.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2VYY
 
 | | Mutant Ala55Trp of Cerebratuls lacteus mini-hemoglobin | | Descriptor: | GLYCEROL, NEURAL HEMOGLOBIN, OXYGEN MOLECULE, ... | | Authors: | Salter, M.D, Nienhaus, K, Nienhaus, G.U, Dewilde, S, Moens, L, Pesce, A, Nardini, M, Bolognesi, M, Olson, J.S. | | Deposit date: | 2008-07-29 | | Release date: | 2008-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Apolar Channel in Cerebratulus Lacteus Hemoglobin is the Route for O2 Entry and Exit.
J.Biol.Chem., 283, 2008
|
|
2VYZ
 
 | | Mutant Ala55Phe of Cerebratulus lacteus mini-hemoglobin | | Descriptor: | ACETATE ION, GLYCEROL, NEURAL HEMOGLOBIN, ... | | Authors: | Salter, M.D, Nienhaus, K, Nienhaus, G.U, Dewilde, S, Moens, L, Pesce, A, Nardini, M, Bolognesi, M, Olson, J.S. | | Deposit date: | 2008-07-29 | | Release date: | 2008-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Apolar Channel in Cerebratulus Lacteus Hemoglobin is the Route for O2 Entry and Exit.
J.Biol.Chem., 283, 2008
|
|
1HL3
 
 | | CtBP/BARS in ternary complex with NAD(H) and PIDLSKK peptide | | Descriptor: | C-TERMINAL BINDING PROTEIN 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PRO-ILE-ASP-LEU-SER-LYS-LYS PEPTIDE | | Authors: | Nardini, M, Spano, S, Cericola, C, Pesce, A, Massaro, A, Millo, E, Luini, A, Corda, D, Bolognesi, M. | | Deposit date: | 2003-03-13 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Ctbp/Bars: A Dual-Function Protein Involved in Transcription Co-Repression and Golgi Membrane Fission
Embo J., 22, 2003
|
|
5MUD
 
 | | Crystal structure of an amyloidogenic light chain dimer H6 | | Descriptor: | light chain dimer,IGL@ protein | | Authors: | Oberti, L, Bacarizo, J, Maritan, M, Rognoni, P, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
