6KZI
 
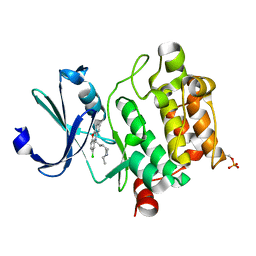 | | Crystal structure of Ser/Thr kinase Pim1 in complex with thioridazine derivatives | | Descriptor: | 4-(2-chloro-10H-phenoxazin-10-yl)-N,N-diethylbutan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Wan, X, Li, W, Xie, Y, Huang, N. | | Deposit date: | 2019-09-24 | | Release date: | 2020-03-04 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
3ILQ
 
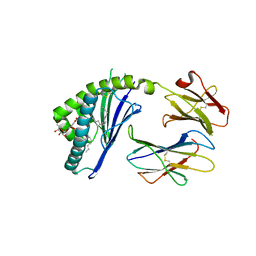 | | Structure of mCD1d with bound glycolipid BbGL-2c from Borrelia burgdorferi | | Descriptor: | (2S)-3-(alpha-D-galactopyranosyloxy)-2-(hexadecanoyloxy)propyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M. | | Deposit date: | 2009-08-07 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ILP
 
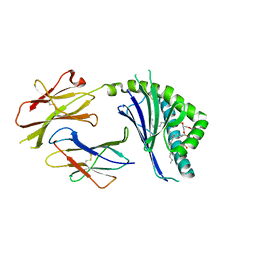 | | Structure of mCD1d with bound glycolipid BbGL-2f from Borrelia burgdorferi | | Descriptor: | (2S)-3-(alpha-D-galactopyranosyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl (9Z,12Z)-octadeca-9,12-dienoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M. | | Deposit date: | 2009-08-07 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8W8Q
 
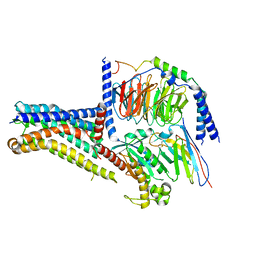 | | Cryo-EM structure of the GPR101-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Gao, N, Yu, X, Wang, G.P, Yang, F, Wang, J.Y, Yang, Z, Guan, Y. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
6A50
 
 | | structure of benzoylformate decarboxylases in complex with cofactor TPP | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, benzoylformate decarboxylases | | Authors: | Guo, Y, Wang, S, Nie, Y, Li, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Synthetic Pathway for Acetyl-Coenzyme A Biosynthesis
Nat Commun, 2019
|
|
8W8S
 
 | | Cryo-EM structure of the AA14-bound GPR101 complex | | Descriptor: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Probable G-protein coupled receptor 101 | | Authors: | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8W8R
 
 | | Cryo-EM structure of the AA-14-bound GPR101-Gs complex | | Descriptor: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
5W0F
 
 | | CREBBP Bromodomain in complex with Cpd3 ((S)-1-(3-(6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydrofuran-3-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-[(3S)-oxolan-3-yl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-30 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
5W0L
 
 | | CREBBP Bromodomain in complex with Cpd10 (1-(3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydro-2H-pyran-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-(oxan-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-31 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
3WJM
 
 | | Crystal structure of Bombyx mori Sp2/Sp3 heterohexamer | | Descriptor: | Arylphorin, Silkworm storage protein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yuan, Y.A, Hou, Y. | | Deposit date: | 2013-10-11 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Bombyx mori arylphorins reveals a 3:3 heterohexamer with multiple papain cleavage sites
Protein Sci., 23, 2014
|
|
5GWN
 
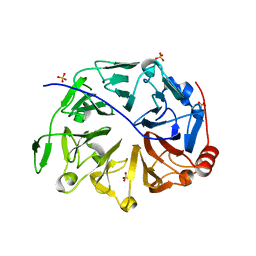 | | Crystal structure of human RCC2 | | Descriptor: | Protein RCC2, SULFATE ION | | Authors: | Liang, L, Yun, C.H, Yin, Y.X. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.309 Å) | | Cite: | RCC2 is a novel p53 target in suppressing metastasis.
Oncogene, 37, 2018
|
|
5W0X
 
 | | Crystal structure of mouse TOR signaling pathway regulator-like (TIPRL) delta 94-103 | | Descriptor: | TIP41-like protein | | Authors: | Wu, C, Zheng, A, Li, J, Satyshur, K, Xing, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-17 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | Methylation-regulated decommissioning of multimeric PP2A complexes.
Nat Commun, 8, 2017
|
|
7XVK
 
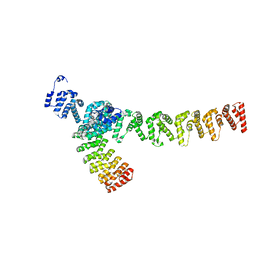 | |
7XVI
 
 | |
4OLS
 
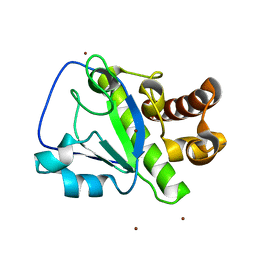 | | The amidase-2 domain of LysGH15 | | Descriptor: | Endolysin, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Gu, J, Ouyang, S, Liu, Z.J, Han, W. | | Deposit date: | 2014-01-24 | | Release date: | 2014-06-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and biochemical characterization reveals LysGH15 as an unprecedented "EF-hand-like" calcium-binding phage lysin.
Plos Pathog., 10, 2014
|
|
5W0W
 
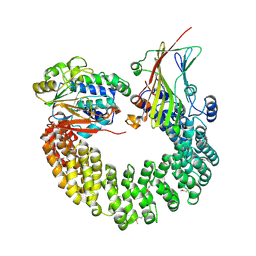 | | Crystal structure of Protein Phosphatase 2A bound to TIPRL | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform, ... | | Authors: | Wu, C, Zheng, A, Li, J, Satyshur, K, Xing, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-17 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Methylation-regulated decommissioning of multimeric PP2A complexes.
Nat Commun, 8, 2017
|
|
4OLK
 
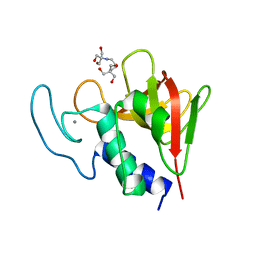 | | The CHAP domain of LysGH15 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Endolysin | | Authors: | Gu, J, Ouyang, S, Liu, Z.J, Han, W. | | Deposit date: | 2014-01-24 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Structural and biochemical characterization reveals LysGH15 as an unprecedented "EF-hand-like" calcium-binding phage lysin.
Plos Pathog., 10, 2014
|
|
5DDT
 
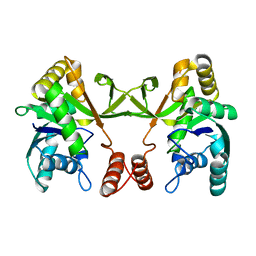 | |
4RXJ
 
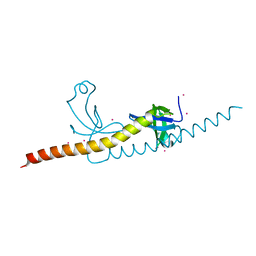 | | crystal structure of WHSC1L1-PWWP2 | | Descriptor: | Histone-lysine N-methyltransferase NSD3, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Dong, A, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-11 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histone and DNA binding ability studies of the NSD subfamily of PWWP domains.
Biochem.Biophys.Res.Commun., 569, 2021
|
|
3LHG
 
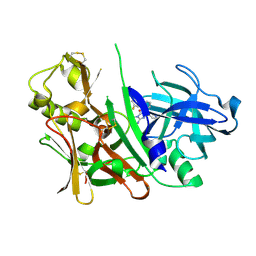 | | Bace1 in complex with the aminohydantoin Compound 4g | | Descriptor: | (5S)-2-amino-5-(2',5'-difluorobiphenyl-3-yl)-3-methyl-5-pyridin-4-yl-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2010-01-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyridinyl aminohydantoins as small molecule BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3T9N
 
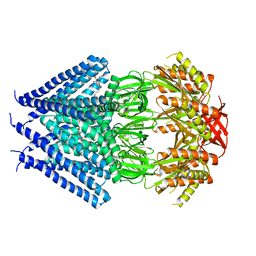 | | Crystal structure of a membrane protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Small-conductance mechanosensitive channel | | Authors: | Yang, M, Zhang, X, Ge, J, Wang, J. | | Deposit date: | 2011-08-03 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.456 Å) | | Cite: | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5D4Y
 
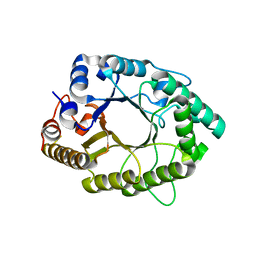 | | A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase | | Descriptor: | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, xylanase | | Authors: | Zheng, Y, Guo, R.T. | | Deposit date: | 2015-08-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into potential cold adaptation mechanism through a psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase.
J.Struct.Biol., 193, 2016
|
|
5DDV
 
 | |
8HR2
 
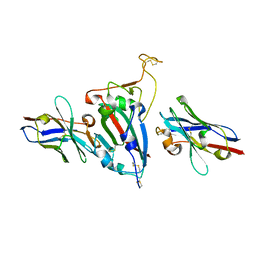 | | Ternary Crystal Complex Structure of RBD with NB1B5 and NB1C6 | | Descriptor: | NB1B5, NB1C6, Spike protein S1 | | Authors: | Sun, Z. | | Deposit date: | 2022-12-14 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure basis of two nanobodies neutralizing SARS-CoV-2 Omicron variant by targeting ultra-conservative epitopes.
J.Struct.Biol., 215, 2023
|
|
7XN4
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with NAD+ | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
