8A8C
 
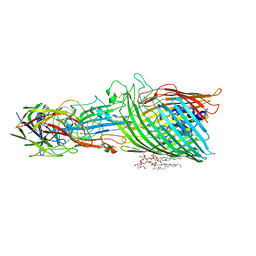 | | T5 phage receptor-binding protein pb5 bound to ferrichrome transporter FhuA | | Descriptor: | Ferrichrome outer membrane transporter/phage receptor, Receptor-binding protein pb5, [(2R,3S,4R,5R,6R)-2-[[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-4-[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-2-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-2-carboxy-5-oxidanyl-oxan-2-yl]oxymethyl]-5-[[(3R)-3-dodecanoyloxytetradecanoyl]amino]-4-(3-nonanoyloxypropanoyloxy)-6-[[(2R,3S,4R,5R,6R)-3-oxidanyl-4-[(3S)-3-oxidanyltetradecanoyl]oxy-5-[[(3R)-3-oxidanyltridecanoyl]amino]-6-phosphonatooxy-oxan-2-yl]methoxy]oxan-3-yl] phosphate | | Authors: | Silale, A, van den Berg, B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for host recognition and superinfection exclusion by bacteriophage T5.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6O66
 
 | |
1N68
 
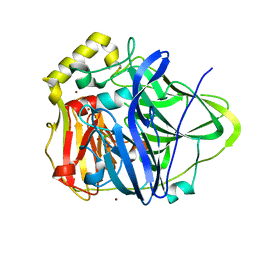 | | Copper bound to the Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-CL-CU LINKAGE | | Authors: | Roberts, S.A, Wildner, G.F, Grass, G, Weichsel, A, Ambrus, A, Rensing, C, Montfort, W.R. | | Deposit date: | 2002-11-08 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Labile Regulatory Copper Ion Lies Near the T1 Copper Site in the Multicopper Oxidase CueO.
J.Biol.Chem., 278, 2003
|
|
6IBP
 
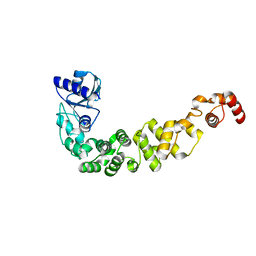 | | Structure of a psychrophilic CCA-adding enzyme at room temperature in ChipX microfluidic device | | Descriptor: | CCA-adding enzyme | | Authors: | de Wijn, R, Hennig, O, Rollet, K, Bluhm, A, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2018-11-30 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.536 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
6OCL
 
 | | Crystal Structure of Leporine Serum Albumin in Complex with Suprofen | | Descriptor: | (2S)-2-[4-(thiophene-2-carbonyl)phenyl]propanoic acid, ACETATE ION, Serum albumin, ... | | Authors: | Zielinski, K, Sekula, B, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-03-24 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural investigations of stereoselective profen binding by equine and leporine serum albumins.
Chirality, 32, 2020
|
|
1GHC
 
 | | HOMO-AND HETERONUCLEAR TWO-DIMENSIONAL NMR STUDIES OF THE GLOBULAR DOMAIN OF HISTONE H1: FULL ASSIGNMENT, TERTIARY STRUCTURE, AND COMPARISON WITH THE GLOBULAR DOMAIN OF HISTONE H5 | | Descriptor: | GH1 | | Authors: | Cerf, C, Lippens, G, Ramakrishnan, V, Muyldermans, S, Segers, A, Wyns, L, Wodak, S.J, Hallenga, K. | | Deposit date: | 1994-05-16 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1: full assignment, tertiary structure, and comparison with the globular domain of histone H5.
Biochemistry, 33, 1994
|
|
2WAC
 
 | |
2V98
 
 | | Structure of the complex of TcAChE with 1-(2-nitrophenyl)-2,2,2- trifluoroethyl-arsenocholine after a 9 seconds annealing to room temperature, during the first 5 seconds of which laser irradiation at 266nm took place | | Descriptor: | 1-(2-nitrophenyl)-2,2,2-trifluoroethyl]-arsenocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Colletier, J.-P, Sanson, B, Royant, A, Specht, A, Nachon, F, Masson, P, Zaccai, G, Sussman, J.L, Goeldner, M, Silman, I, Bourgeois, D, Weik, M. | | Deposit date: | 2007-08-22 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Use of a 'Caged' Analog to Study Traffic of Choline within Acetylcholinesterase by Kinetic Crystallography
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6OUJ
 
 | | Carbonic Anhydrase II complexed with benzene sulfonamide MB11-689A | | Descriptor: | 3-[(1S)-1-(4-nitrophenyl)ethyl]-2-oxo-2,3-dihydro-1,3-benzoxazole-5-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Kota, A, McKenna, R. | | Deposit date: | 2019-05-04 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.466 Å) | | Cite: | Carbonic Anhydrase II complexed with benzene sulfonamide MB11-689A
To Be Published
|
|
4LDV
 
 | | Crystal structure of the DNA binding domain of A. thailana auxin response factor 1 | | Descriptor: | Auxin response factor 1, CHLORIDE ION, FORMIC ACID, ... | | Authors: | boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
6OXX
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-18 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1R)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3 | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
2VMD
 
 | | Structure of the complex of discoidin II from Dictyostelium discoideum with beta-methyl-galactose | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CALCIUM ION, ... | | Authors: | Aragao, K.S, Satre, M, Imberty, A, Varrot, A. | | Deposit date: | 2008-01-25 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure Determination of Discoidin II from Dictyostelium Discoideum and Carbohydrate Binding Properties of the Lectin Domain.
Proteins, 73, 2008
|
|
6FZR
 
 | | Crystal structure of scFv-SM3 in complex with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-[(fluoroacetyl)amino]-alpha-D-galactopyranose, Mucin-1, ... | | Authors: | Bermejo, I.A, Usabiaga, I, Companon, I, Castro-Lopez, J, Insausti, A, Fernandez, J.A, Avenoza, A, Busto, J.H, Jimenez-Barbero, J, Asensio, J.L, Jimenez-Oses, G, Peregrina, J.M, Hurtado-Guerrero, R, Cocinero, E.J, Corzana, F. | | Deposit date: | 2018-03-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Water Sculpts the Distinctive Shapes and Dynamics of the Tumor-Associated Carbohydrate Tn Antigens: Implications for Their Molecular Recognition.
J.Am.Chem.Soc., 140, 2018
|
|
1G82
 
 | | STRUCTURE OF FIBROBLAST GROWTH FACTOR 9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, FIBROBLAST GROWTH FACTOR 9, ... | | Authors: | Hecht, H.J, Adar, R, Hofmann, B, Bogin, O, Weich, H, Yayon, A. | | Deposit date: | 2000-11-16 | | Release date: | 2001-03-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of fibroblast growth factor 9 shows a symmetric dimer with unique receptor- and heparin-binding interfaces.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6OTO
 
 | |
4YD7
 
 | | Endothiapepsin in complex with fragment 255 | | Descriptor: | 2-(imidazo[1,2-a]pyridin-2-yl)-N-phenylacetamide, ACETATE ION, Endothiapepsin, ... | | Authors: | Stieler, M, Heine, A, Klebe, G. | | Deposit date: | 2015-02-21 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
6Q4Y
 
 | | Structure of MPT-2, a GDP-Man-dependent mannosyltransferase from Leishmania mexicana, in complex with mannose | | Descriptor: | LmxM MPT-2, alpha-D-mannopyranose, beta-D-mannopyranose | | Authors: | Sobala, L.F, Males, A, Bastidas, L.M, Ward, T, Sernee, M.F, Ralton, J.E, Nero, T.L, Cobbold, S, Kloehn, J, Viera-Lara, M, Stanton, L, Hanssen, E, Parker, M.W, Williams, S.J, McConville, M.J, Davies, G.J. | | Deposit date: | 2018-12-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Family of Dual-Activity Glycosyltransferase-Phosphorylases Mediates Mannogen Turnover and Virulence in Leishmania Parasites.
Cell Host Microbe, 26, 2019
|
|
6Q7F
 
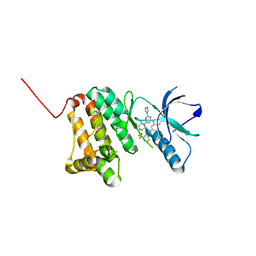 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with the NVP-BHG712 derivative ATDL18 | | Descriptor: | 3-[(4-imidazol-1-yl-6-piperazin-1-yl-1,3,5-triazin-2-yl)amino]-4-methyl-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Gande, S.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-12-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.204 Å) | | Cite: | Effects of NVP-BHG712 chemical modifications on EPHA2 binding and affinity
To Be Published
|
|
2V3Q
 
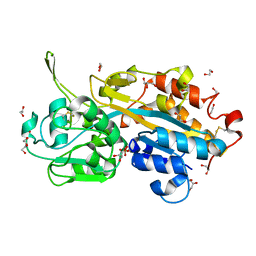 | | Serendipitous discovery and X-ray structure of a human phosphate binding apolipoprotein | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, HUMAN PHOSPHATE BINDING PROTEIN, ... | | Authors: | Morales, R, Berna, A, Carpentier, P, Elias, M, Contreras-Martel, C, Renault, F, Nicodeme, M, Chesne-Seck, M.-L, Bernier, F, Dupuy, J, Schaeffer, C, Diemer, H, Van Dorsselaer, A, Fontecilla, J.C, Masson, P, Rochu, D, Chabriere, E. | | Deposit date: | 2007-06-20 | | Release date: | 2008-07-22 | | Last modified: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Tandem Use of X-Ray Crystallography and Mass Spectrometry to Obtain Ab Initio the Complete and Exact Amino Acids Sequence of Hpbp, a Human 38kDa Apolipoprotein
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
7ZZK
 
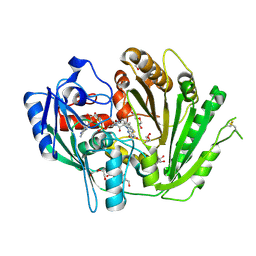 | | Structure of the N-acetyl-D-glucosamine oxidase from Ralstonia Solanacearum | | Descriptor: | ALANINE, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Boverio, A, Rozeboom, H.J, Fraaije, M.W. | | Deposit date: | 2022-05-25 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Elucidation and Engineering of a Bacterial Carbohydrate Oxidase.
Biochemistry, 62, 2023
|
|
7ZOT
 
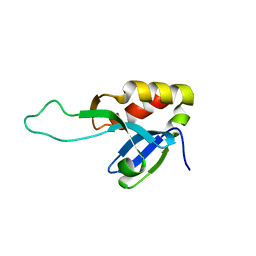 | |
6JDW
 
 | |
6QA1
 
 | | ERK2 mini-fragment binding | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, pyridin-2-amine | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
6GA8
 
 | | BACTERIORHODOPSIN, 330 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6G7J
 
 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 457-646 fs state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
