8WRH
 
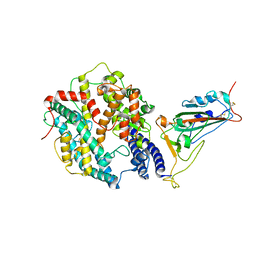 | | SARS-CoV-2 XBB.1.5.70 in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
4QR5
 
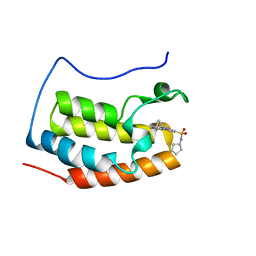 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | Bromodomain-containing protein 4, N-[3-(cyclopentylsulfamoyl)-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]cyclopropanecarboxamide | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
8WRL
 
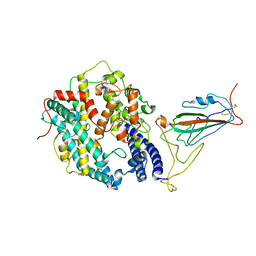 | | XBB.1.5 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
4DS8
 
 | | Complex structure of abscisic acid receptor PYL3-(+)-ABA-HAB1 in the presence of Mn2+ | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, GLYCEROL, ... | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
8WRM
 
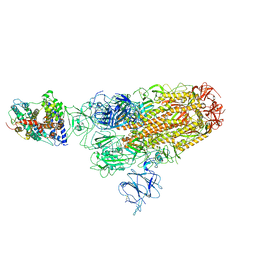 | | XBB.1.5 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRO
 
 | | XBB.1.5.10 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein,Spike glycoprotein,Spike glycoprotein,Fusion protein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTD
 
 | | XBB.1.5.10 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTJ
 
 | | XBB.1.5.70 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.64 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
4LUQ
 
 | |
4JGV
 
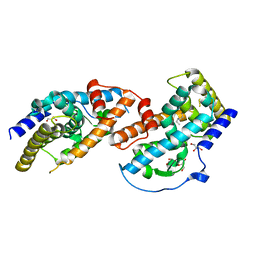 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with THPN | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Li, F, Li, A, Tian, X, Wan, W, Wan, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-03-04 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
6NZT
 
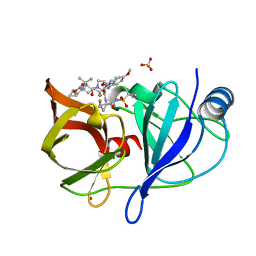 | |
6KC0
 
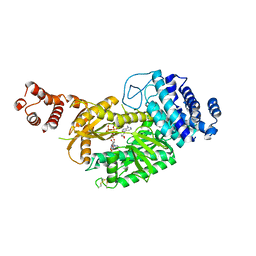 | | fused To-MtbCsm1 with 2ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A),CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), MAGNESIUM ION | | Authors: | Li, T, Huo, Y, Jiang, T. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Mycobacterium tuberculosis CRISPR/Cas system Csm1 holds clues to the evolutionary relationship between DNA polymerase and cyclase activity.
Int.J.Biol.Macromol., 170, 2020
|
|
6U8X
 
 | | Crystal structure of DNMT3B-DNMT3L in complex with CpApG DNA | | Descriptor: | CpApG DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Song, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95063877 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
7MXN
 
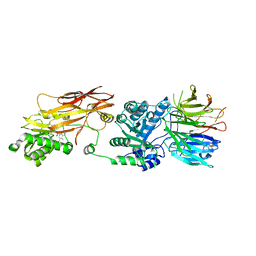 | | PRMT5(M420T mutant):MEP50 complexed with inhibitor PF-06939999 | | Descriptor: | (1S,2S,3S,5R)-3-{[6-(difluoromethyl)-5-fluoro-1,2,3,4-tetrahydroisoquinolin-8-yl]oxy}-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclopentane-1,2-diol, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
7MX7
 
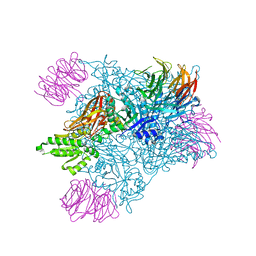 | | PRMT5:MEP50 complexed with inhibitor PF-06939999 | | Descriptor: | (1S,2S,3S,5R)-3-{[6-(difluoromethyl)-5-fluoro-1,2,3,4-tetrahydroisoquinolin-8-yl]oxy}-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclopentane-1,2-diol, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
7MXA
 
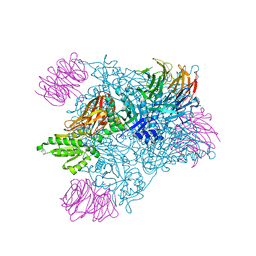 | | PRMT5:MEP50 complexed with inhibitor PF-06855800 | | Descriptor: | 7-[(5R)-5-C-(4-chloro-3-fluorophenyl)-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.713 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
7MXC
 
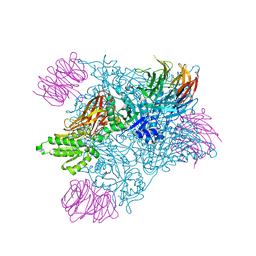 | | PRMT5:MEP50 complexed with adenosine | | Descriptor: | ADENOSINE, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
7MXG
 
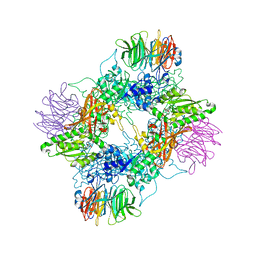 | | PRMT5(M420T mutant):MEP50 complexed with inhibitor PF-06855800 | | Descriptor: | 7-[(5R)-5-C-(4-chloro-3-fluorophenyl)-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
6U8P
 
 | | Crystal structure of DNMT3B-DNMT3L in complex with CpGpA DNA | | Descriptor: | CpGpA DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Song, J. | | Deposit date: | 2019-09-05 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
6U8V
 
 | | Crystal structure of DNMT3B-DNMT3L in complex with CpGpT DNA | | Descriptor: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Zhang, Z.M, Song, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
6U8W
 
 | | Crystal structure of DNMT3B(K777A)-DNMT3L in complex with CpGpT DNA | | Descriptor: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Zhang, Z.M, Song, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.94891548 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
4DSC
 
 | | Complex structure of abscisic acid receptor PYL3 with (+)-ABA in spacegroup of H32 at 1.95A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION | | Authors: | Zhang, X, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
4X9Z
 
 | | Dimeric conotoxin alphaD-GeXXA | | Descriptor: | alphaD-conotoxin GeXXA from the venom of Conus generalis | | Authors: | Xu, S, Zhang, T, Kompella, S, Adams, D, Ding, J, Wang, C. | | Deposit date: | 2014-12-12 | | Release date: | 2015-12-02 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conotoxin alpha D-GeXXA utilizes a novel strategy to antagonize nicotinic acetylcholine receptors
Sci Rep, 5, 2015
|
|
8GQE
 
 | | Crystal structure of the W285A mutant of UVR8 in complex with RUP2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ultraviolet-B receptor UVR8, WD repeat-containing protein RUP2 | | Authors: | Wang, Y.D, Wang, L.X, Guan, Z.Y, chang, H.F, Yin, P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RUP2 facilitates UVR8 redimerization via two interfaces.
Plant Commun., 4, 2023
|
|
