1OHE
 
 | | Structure of cdc14b phosphatase with a peptide ligand | | Descriptor: | CDC14B2 PHOSPHATASE, PEPTIDE LIGAND | | Authors: | Gray, C.H, Good, V.M, Tonks, N.K, Barford, D. | | Deposit date: | 2003-05-24 | | Release date: | 2003-07-24 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Cell Cycle Protein Cdc14 Reveals a Proline-Directed Protein Phosphatase
Embo J., 22, 2003
|
|
1OPK
 
 | | Structural basis for the auto-inhibition of c-Abl tyrosine kinase | | Descriptor: | 6-(2,6-DICHLOROPHENYL)-2-{[3-(HYDROXYMETHYL)PHENYL]AMINO}-8-METHYLPYRIDO[2,3-D]PYRIMIDIN-7(8H)-ONE, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Nagar, B, Hantschel, O, Young, M.A, Scheffzek, K, Veach, D, Bornmann, W, Clarkson, B, Superti-Furga, G, Kuriyan, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the autoinhibition of c-Abl tyrosine kinase
Cell(Cambridge,Mass.), 112, 2003
|
|
1OKN
 
 | | CARBONIC ANHYDRASE II COMPLEX WITH THE 1OKN INHIBITOR 4-SULFONAMIDE-[1-(4-N-(5-FLUORESCEIN THIOUREA)BUTANE)] | | Descriptor: | 4-SULFONAMIDE-[4-(THIOMETHYLAMINOBUTANE)]BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Elbaum, D, Nair, S.K, Patchan, M.W, Thompson, R.B, Christianson, D.W. | | Deposit date: | 1996-06-25 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of a sulfonamide probe for fluorescence anisotropy detection of zinc with a carbonic anhydrase-based biosensor.
J.Am.Chem.Soc., 118, 1996
|
|
1OG7
 
 | | Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P. | | Descriptor: | BACTERIOCIN SAKACIN P | | Authors: | Uteng, M, Hauge, H.H, Markwick, P.R, Fimland, G, Mantzilas, D, Nissen-Meyer, J, Muhle-Goll, C. | | Deposit date: | 2003-04-25 | | Release date: | 2003-09-22 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure in Lipid Micelles of the Pediocin-Like Antimicrobial Peptide Sakacin P and a Sakacin P Variant that is Structurally Stabilized by an Inserted C-Terminal Disulfide Bridge
Biochemistry, 42, 2003
|
|
1OGQ
 
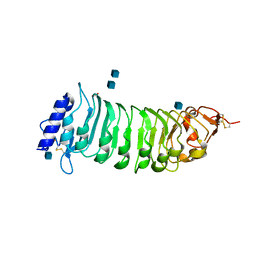 | | The crystal structure of PGIP (polygalacturonase inhibiting protein), a leucine rich repeat protein involved in plant defense | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, POLYGALACTURONASE INHIBITING PROTEIN | | Authors: | Di Matteo, A, Federici, L, Mattei, B, Salvi, G, Johnson, K.A, Savino, C, De Lorenzo, G, Tsernoglou, D, Cervone, F. | | Deposit date: | 2003-05-08 | | Release date: | 2003-07-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Polygalacturonase-Inhibiting Protein (Pgip), a Leucine-Rich Repeat Protein Involved in Plant Defense
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7OV8
 
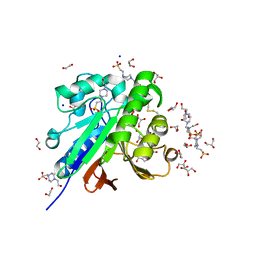 | | Crystal structure of pig purple acid phosphatase in complex with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) and glycerol | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Feder, D, McGeary, R.P, Guddat, L.W, Schenk, G. | | Deposit date: | 2021-06-14 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design of Potent Inhibitors of a Metallohydrolase Using a Fragment-Based Approach.
Chemmedchem, 16, 2021
|
|
1OHN
 
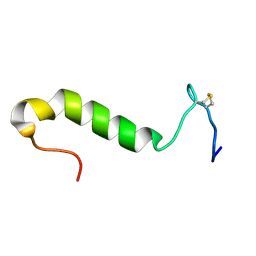 | | Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P. | | Descriptor: | BACTERIOCIN SAKACIN P | | Authors: | Uteng, M, Hauge, H.H, Markwick, P.R, Fimland, G, Mantzilas, D, Nissen-Meyer, J, Muhle-Goll, C. | | Deposit date: | 2003-05-28 | | Release date: | 2003-09-22 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure in Lipid Micelles of the Pediocin-Like Antimicrobial Peptide Sakacin P and a Sakacin P Variant that is Structurally Stabilized by an Inserted C-Terminal Disulfide Bridge
Biochemistry, 42, 2003
|
|
5MWO
 
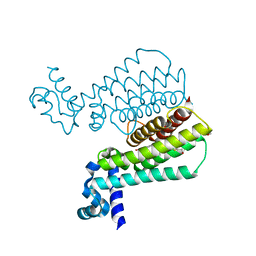 | | Structure of Mycobacterium Tuberculosis Transcriptional Regulatory Repressor Protein (EthR) in complex with fragment 7E8. | | Descriptor: | (5-methyl-1-benzothiophen-2-yl)methanol, 1,2-ETHANEDIOL, HTH-type transcriptional regulator EthR | | Authors: | Mendes, V, Chan, D.S.-H, Thomas, S.E, McConnell, B, Matak-Vinkovic, D, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Fragment Screening against the EthR-DNA Interaction by Native Mass Spectrometry.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2UYU
 
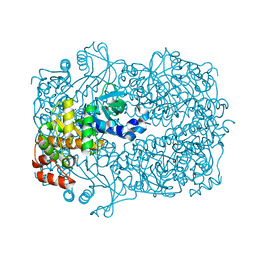 | |
1OH4
 
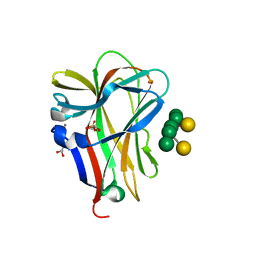 | | Structural and thermodynamic dissection of specific mannan recognition by a carbohydrate-binding module | | Descriptor: | BETA-MANNOSIDASE, CALCIUM ION, GLYCEROL, ... | | Authors: | Boraston, A.B, Revett, T.J, Boraston, C.M, Nurizzo, D, Davies, G.J. | | Deposit date: | 2003-05-21 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Thermodynamic Dissection of Specific Mannan Recognition by a Carbohydrate Binding Module, Tmcbm27
Structure, 11, 2003
|
|
1Q5J
 
 | | Crystal structure of bacteriorhodopsin mutant P91A crystallized from bicelles | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Yohannan, S, Faham, S, Yang, D, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-08-07 | | Release date: | 2004-01-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The evolution of transmembrane helix kinks and the structural diversity of G protein-coupled receptors.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5LG7
 
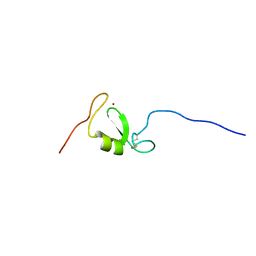 | | Solution NMR structure of Tryptophan to Arginine mutant of Arkadia RING domain | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Birkou, M, Chasapis, C.T, Loutsidou, A.K, Bentrop, D, Lelli, M, Herrmann, T, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2016-07-06 | | Release date: | 2017-06-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Residue Specific Insight into the Arkadia E3 Ubiquitin Ligase Activity and Conformational Plasticity.
J. Mol. Biol., 429, 2017
|
|
2V30
 
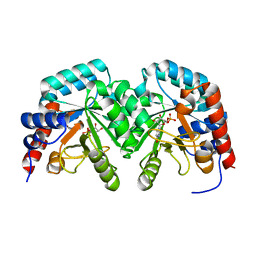 | | Human orotidine 5'-phosphate decarboxylase domain of uridine monophospate synthetase (UMPS) in complex with its product UMP. | | Descriptor: | OROTIDINE 5'-PHOSPHATE DECARBOXYLASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Moche, M, Ogg, D, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, van den Berg, S, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Orotidine 5'-Decarboxylase Domain of Human Uridine Monophosphate Synthetase (Umps)
To be Published
|
|
1QXC
 
 | | NMR structure of the fragment 25-35 of beta amyloid peptide in 20/80 v:v hexafluoroisopropanol/water mixture | | Descriptor: | 11-mer peptide from Amyloid beta A4 protein | | Authors: | D'Ursi, A.M, Armenante, M.R, Guerrini, R, Salvadori, S, Sorrentino, G, Picone, D. | | Deposit date: | 2003-09-05 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide (25-35) in different media
J.Med.Chem., 47, 2004
|
|
1PQM
 
 | | T4 Lysozyme Core Repacking Mutant V149I/T152V/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Lysozyme, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-06-18 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Repacking the Core of T4 Lysozyme by Automated Design
J.Mol.Biol., 332, 2003
|
|
5LKU
 
 | | Crystal structure of the p300 acetyltransferase catalytic core with coenzyme A. | | Descriptor: | COENZYME A, Histone acetyltransferase p300,Histone acetyltransferase p300, ZINC ION | | Authors: | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
5LPW
 
 | |
2VLN
 
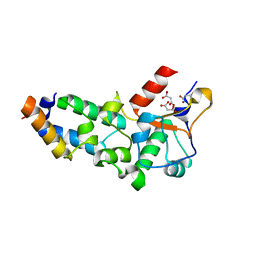 | | N75A mutant of E9 DNase domain in complex with Im9 | | Descriptor: | COLICIN E9, COLICIN-E9 IMMUNITY PROTEIN, MALONIC ACID | | Authors: | Keeble, A.H, Joachimiak, L.A, Mate, M.J, Meenan, N, Kirkpatrick, N, Baker, D, Kleanthous, C. | | Deposit date: | 2008-01-15 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental and Computational Analyses of the Energetic Basis for Dual Recognition of Immunity Proteins by Colicin Endonucleases.
J.Mol.Biol., 379, 2008
|
|
1PMM
 
 | | Crystal structure of Escherichia coli GadB (low pH) | | Descriptor: | ACETIC ACID, Glutamate decarboxylase beta, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Capitani, G, De Biase, D, Aurizi, C, Gut, H, Bossa, F, Grutter, M.G. | | Deposit date: | 2003-06-11 | | Release date: | 2004-02-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and functional analysis of escherichia coli glutamate
decarboxylase
Embo J., 22, 2003
|
|
2V9O
 
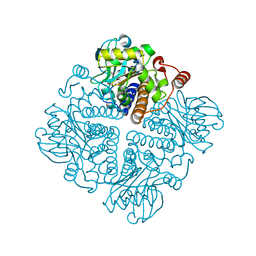 | |
1Q3V
 
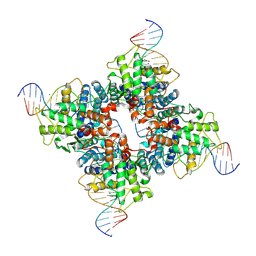 | | Crystal structure of a wild-type Cre recombinase-loxP synapse: phosphotyrosine covalent intermediate | | Descriptor: | Cre recombinase, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Meyer, J.E.W, Buchholz, F, Stewart, A.F, Suck, D. | | Deposit date: | 2003-08-01 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of a wild-type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation
Nucleic Acids Res., 31, 2003
|
|
5LRW
 
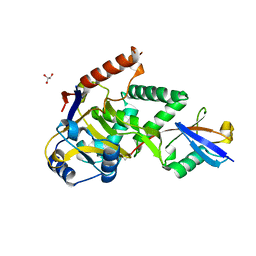 | | Structure of Cezanne/OTUD7B OTU domain bound to ubiquitin | | Descriptor: | GLYCEROL, OTU domain-containing protein 7B, Polyubiquitin-B | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
1Q4T
 
 | | crystal structure of 4-hydroxybenzoyl CoA thioesterase from Arthrobacter sp. strain SU complexed with 4-hydroxyphenyl CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYPHENACYL COENZYME A, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Zhuang, Z, Dunaway-Mariano, D, Holden, H.M. | | Deposit date: | 2003-08-04 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. strain SU
J.Biol.Chem., 278, 2003
|
|
5LSU
 
 | |
5LP6
 
 | | Crystal structure of Tubulin-Stathmin-TTL-Thiocolchicine Complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Marangon, J, Christodoulou, M, Casagrande, F, Tiana, G, Dalla Via, L, Aliverti, A, Passarella, D, Cappelletti, G, Ricagno, S. | | Deposit date: | 2016-08-11 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Tools for the rational design of bivalent microtubule-targeting drugs.
Biochem. Biophys. Res. Commun., 479, 2016
|
|
