6RS6
 
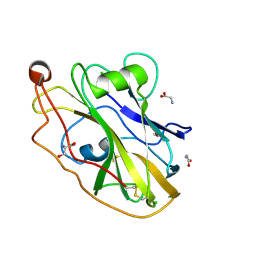 | | X-ray crystal structure of LsAA9B | | Descriptor: | AA9, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Frandsen, K.E.H, Tovborg, M, Poulsen, J.C.N, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2019-05-21 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into an unusual Auxiliary Activity 9 family member lacking the histidine brace motif of lytic polysaccharide monooxygenases.
J.Biol.Chem., 294, 2019
|
|
7YNK
 
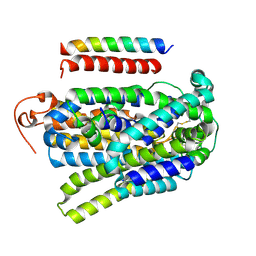 | |
7YPM
 
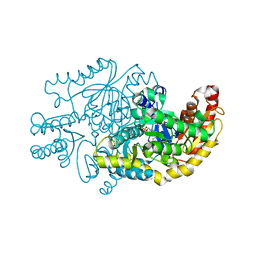 | | Crystal structure of transaminase CC1012 complexed with PLP and L-alanine | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Aspartate aminotransferase family protein, ... | | Authors: | Yang, L, Wang, H, Wei, D. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Mechanism-Guided Computational Design of omega-Transaminase by Reprograming of High-Energy-Barrier Steps.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7YNJ
 
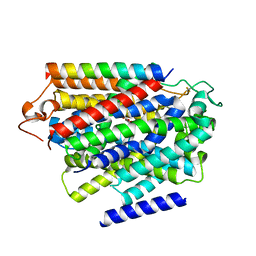 | |
7YNI
 
 | | Structure of human SGLT1-MAP17 complex bound with substrate 4D4FDG in the occluded conformation | | Descriptor: | (2R,3R,4R,5S,6R)-5-fluoranyl-6-(hydroxymethyl)oxane-2,3,4-triol, PDZK1-interacting protein 1, Sodium/glucose cotransporter 1 | | Authors: | Chen, L, Niu, Y, Cui, W. | | Deposit date: | 2022-07-31 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structures of human SGLT in the occluded state reveal conformational changes during sugar transport.
Nat Commun, 14, 2023
|
|
6RZU
 
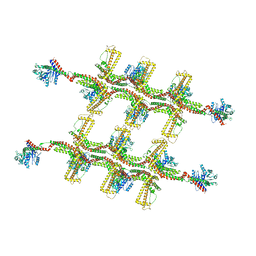 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
8CEE
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CED
 
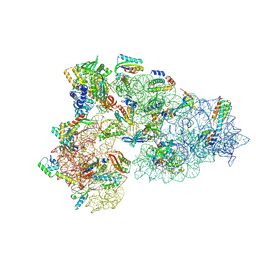 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CDU
 
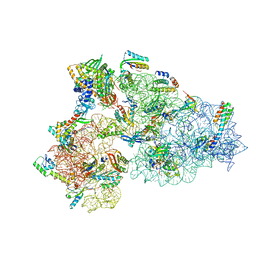 | | Rnase R bound to a 30S degradation intermediate (main state) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
6RFX
 
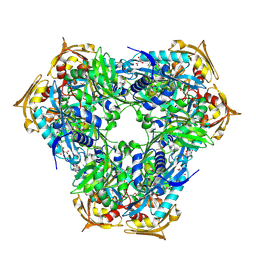 | | Crystal structure of Eis2 from Mycobacterium abscessus | | Descriptor: | ACETATE ION, CITRIC ACID, Eis2, ... | | Authors: | Blaise, M, Kremer, L, Olieric, V, Alsarraf, H, Ung, K.L. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the aminoglycosides N-acetyltransferase Eis2 from Mycobacterium abscessus.
Febs J., 286, 2019
|
|
8CEC
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CDV
 
 | | Rnase R bound to a 30S degradation intermediate (state II) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.73 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
6RW2
 
 | | Bicycle Toxin Conjugate bound to EphA2 | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, ALA-ARG-ASP-CYS-PRO-LEU-VAL-ASN-PRO-LEU-CYS-LEU-HIS-PRO-GLY-TRP-THR-CYS, Ephrin type-A receptor 2, ... | | Authors: | Brown, D.G, Schroeder, S, Chen, L. | | Deposit date: | 2019-06-03 | | Release date: | 2020-04-08 | | Last modified: | 2020-05-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Identification and Optimization of EphA2-Selective Bicycles for the Delivery of Cytotoxic Payloads.
J.Med.Chem., 63, 2020
|
|
6RMQ
 
 | | Crystal structure of a selenomethionine-substituted A70M I84M mutant of the essential repressor DdrO from radiation resistant-Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-07 | | Release date: | 2019-10-09 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
6RY4
 
 | |
6RYU
 
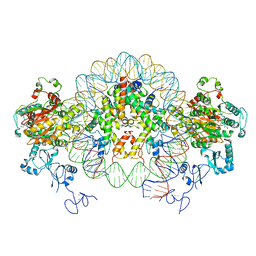 | | Nucleosome-CHD4 complex structure (two CHD4 copies) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4,CHD4,Chromodomain-helicase-DNA-binding protein 4, DNA (149-MER), Histone H2A type 1, ... | | Authors: | Farnung, L, Ochmann, M, Cramer, P. | | Deposit date: | 2019-06-12 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Nucleosome-CHD4 chromatin remodeller structure maps human disease mutations.
Elife, 9, 2020
|
|
6S0G
 
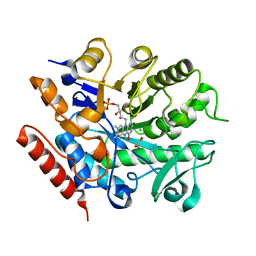 | | Crystal structure of ene-reductase GsOYE from Galdieria sulphuraria | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Robescu, M.R, Niero, M, Hall, M, Bergantino, E, Cendron, L. | | Deposit date: | 2019-06-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Two new ene-reductases from photosynthetic extremophiles enlarge the panel of old yellow enzymes: CtOYE and GsOYE.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
6RV3
 
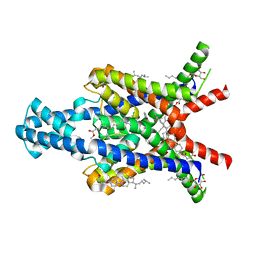 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 1000493 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
8C27
 
 | |
8C5L
 
 | |
6RZT
 
 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RFY
 
 | | Crystal structure of Eis2 form Mycobacterium abscessus | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Eis2, SULFATE ION | | Authors: | Blaise, M, Kremer, L, Olieric, V, Alsarraf, H, Ung, K.L. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the aminoglycosides N-acetyltransferase Eis2 from Mycobacterium abscessus.
Febs J., 286, 2019
|
|
8CQI
 
 | | Human heparanase in complex with inhibitor R3794 | | Descriptor: | (3~{S},4~{S})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, 1,5-anhydro-D-arabinitol, ... | | Authors: | Moran, E.M, Davies, G.J, Chen, C, Nieuwendijk, E.V, Wu, L, Skoulikopoulou, F, Riet, V.V, Overkleeft, H.S, Armstrong, Z. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
6S30
 
 | | Crystal Structure of Seryl-tRNA Synthetase from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Serine--tRNA ligase | | Authors: | Pang, L, De Graef, S, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
8C1J
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_18-30 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Protein arginine N-methyltransferase 2, RNA-binding protein Rsf1-like | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_18-30
To Be Published
|
|
