6ERP
 
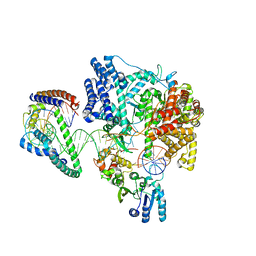 | | Structure of the human mitochondrial transcription initiation complex at the LSP promoter | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, Dimethyladenosine transferase 2, ... | | Authors: | Hillen, H.S, Morozov, Y.I, Sarfallah, A, Temiakov, D, Cramer, P. | | Deposit date: | 2017-10-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.502 Å) | | Cite: | Structural Basis of Mitochondrial Transcription Initiation.
Cell, 171, 2017
|
|
4CO1
 
 | |
4COL
 
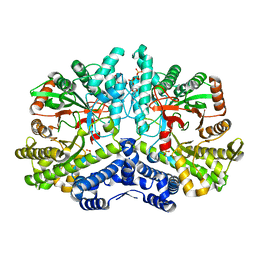 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with dATP bound in the specificity site | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, MAGNESIUM ION, ... | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-29 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
4CVA
 
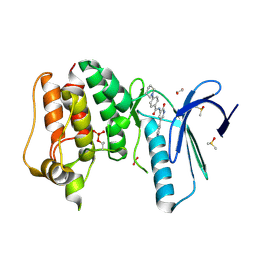 | | MPS1 kinase with 3-aminopyridin-2-one inhibitors | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DUAL SPECIFICITY PROTEIN KINASE TTK, ... | | Authors: | Fearon, D, Bavetsias, V, Bayliss, R, Schmitt, J, Westwood, I.M, vanMontfort, R.L.M, Jones, K. | | Deposit date: | 2014-03-24 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Kinase Selectivity of a 3-Aminopyridin-2- One Based Fragment Library, Identification of 3-Amino-5-(Pyridin-4-Yl)Pyridin-2(1H)-One as a Novel Scaffold for Mps1 Inhibition
To be Published
|
|
4CW9
 
 | | Entamoeba histolytica thiredoxin C34S mutant | | Descriptor: | THIOREDOXIN | | Authors: | Parsonage, D, Kells, P.M, Vieira, D.F, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2014-04-01 | | Release date: | 2015-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
6E1L
 
 | | GRN3Ala | | Descriptor: | Granulin | | Authors: | Dastpeyman, M, Bansal, P, Wilson, D, Sotillo, J, Brindley, P, Loukas, A, Smout, M, Daly, N. | | Deposit date: | 2018-07-10 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structural Variants of a Liver Fluke Derived Granulin Peptide Potently Stimulate Wound Healing.
J. Med. Chem., 61, 2018
|
|
4D1C
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH bromovinylhydantoin bound. | | Descriptor: | (5Z)-5-[(3-bromophenyl)methylidene]imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
6E0W
 
 | | Crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 complexed with one repeating unit of colanic acid | | Descriptor: | 1,2-ETHANEDIOL, 4,6-O-[(1R)-1-carboxyethylidene]-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-alpha-D-galactopyranose-(1-3)-alpha-L-fucopyranose-(1-4)-alpha-L-fucopyranose-(1-3)-beta-D-glucopyranose, Bacteriophage Phi92 gp150, ... | | Authors: | Plattner, M, Browning, C, Gerardy-Schahn, R, Shneider, M.M, Leiman, P.G, Schwarzer, D. | | Deposit date: | 2018-07-07 | | Release date: | 2019-07-17 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 complexed with one repeating unit of colanic acid
To Be Published
|
|
4D55
 
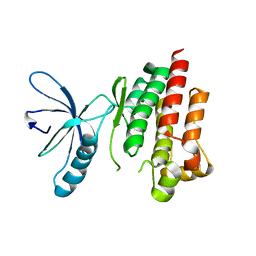 | |
4D4S
 
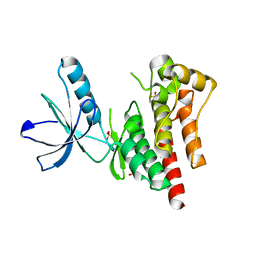 | | Focal Adhesion Kinase catalytic domain | | Descriptor: | 2-({5-CHLORO-2-[(2-METHOXY-4-MORPHOLIN-4-YLPHENYL)AMINO]PYRIMIDIN-4-YL}AMINO)-N-METHYLBENZAMIDE, DIMETHYL SULFOXIDE, FOCAL ADHESION KINASE 1, ... | | Authors: | Le Coq, J, Lin, A, Lietha, D. | | Deposit date: | 2014-10-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric Regulation of Focal Adhesion Kinase by Pip2 and ATP.
Biophys.J., 108, 2015
|
|
6EY9
 
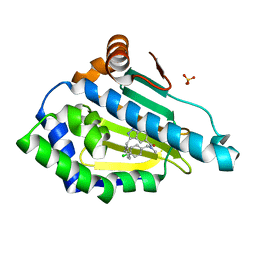 | |
6EZH
 
 | | Torpedo californica AChE in complex with indolic multi-target directed ligand | | Descriptor: | 1-(7-chloranyl-4-methoxy-1~{H}-indol-5-yl)-3-[1-(cyclohexylmethyl)piperidin-4-yl]propan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Santoni, G, Lalut, J, Karila, D, Lecoutey, C, Davis, A, Nachon, F, Sillman, I, Sussman, J, Weik, M, Maurice, T, Dallemagne, P, Rochais, C. | | Deposit date: | 2017-11-15 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel multitarget-directed ligands targeting acetylcholinesterase and sigma1receptors as lead compounds for treatment of Alzheimer's disease: Synthesis, evaluation, and structural characterization of their complexes with acetylcholinesterase.
Eur J Med Chem, 162, 2018
|
|
4CJ7
 
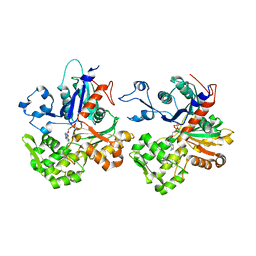 | | Structure of Crenactin, an archeal actin-like protein | | Descriptor: | ACTIN/ACTIN FAMILY PROTEIN, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Izore, T, Duman, R.E, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crenactin from Pyrobaculum Calidifontis is Closely Related to Actin in Structure and Forms Steep Helical Filaments.
FEBS Lett., 588, 2014
|
|
4CPE
 
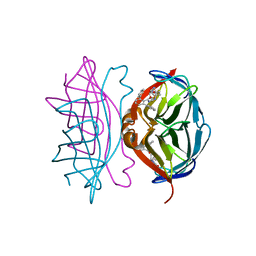 | | Wild-type streptavidin in complex with love-hate ligand 1 (LH1) | | Descriptor: | (3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4- d]imidazolidin-4-yl]-N-{2-[(2,6- diphenylphenyl)formamido]ethyl}pentanamide, STREPTAVIDIN | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
6EYR
 
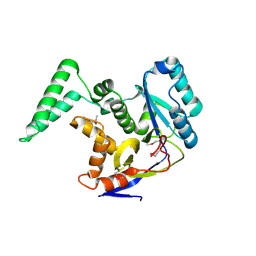 | |
4CYE
 
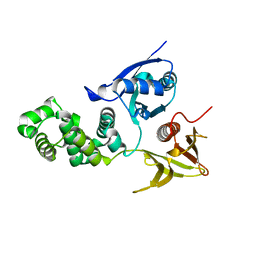 | | Crystal structure of avian FAK FERM domain FAK31-405 at 3.2A | | Descriptor: | FOCAL ADHESION KINASE 1 | | Authors: | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | Deposit date: | 2014-04-10 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6EZY
 
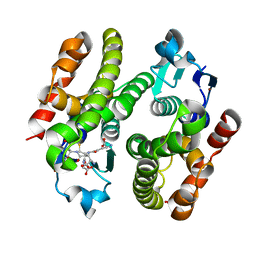 | | ARABIDOPSIS THALIANA GSTF9, GSH AND GSOH BOUND | | Descriptor: | BROMIDE ION, GLUTATHIONE, GLYCEROL, ... | | Authors: | Tossounian, M.A, Wahni, K, VanMolle, I, Rosado, L, Vertommen, D, Messens, J. | | Deposit date: | 2017-11-16 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Redox-regulated methionine oxidation of Arabidopsis thaliana glutathione transferase Phi9 induces H-site flexibility.
Protein Sci., 28, 2019
|
|
4D4G
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | APNAA1, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D4R
 
 | | Focal Adhesion Kinase catalytic domain | | Descriptor: | FOCAL ADHESION KINASE 1, SULFATE ION | | Authors: | Le Coq, J, Lin, A, Lietha, D. | | Deposit date: | 2014-10-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Allosteric Regulation of Focal Adhesion Kinase by Pip2 and ATP.
Biophys.J., 108, 2015
|
|
4D57
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, APNA A1, ARGININE, ... | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-11-03 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D7S
 
 | | Structure of the SthK Carboxy-Terminal Region in complex with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, STHK_CNBD_CGMP | | Authors: | Kesters, D, Brams, M, Nys, M, Wijckmans, E, Spurny, R, Voets, T, Tytgat, J, Ulens, C. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the SthK Carboxy-Terminal Region Reveals a Gating Mechanism for Cyclic Nucleotide-Modulated Ion Channels.
Plos One, 10, 2015
|
|
4D7N
 
 | | TetR(D) in complex with anhydrotetracycline and potassium | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Werten, S, Dalm, D, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-11-25 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Tetracycline Repressor Allostery Does not Depend on Divalent Metal Recognition.
Biochemistry, 53, 2014
|
|
4CNY
 
 | |
4CO2
 
 | |
4D4I
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | APNAA1, ARGININE, GLYCEROL, ... | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
