2R6M
 
 | | Crystal structure of rat CK2-beta subunit | | Descriptor: | Casein kinase II subunit beta, ZINC ION | | Authors: | Shen, Y. | | Deposit date: | 2007-09-06 | | Release date: | 2008-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of catalytic and regulatory subunits of rat protein kinase CK2
CHIN.SCI.BULL., 54, 2009
|
|
4AZA
 
 | | Improved eIF4E binding peptides by phage display guided design. | | Descriptor: | EIF4G1_D5S PEPTIDE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, [[(2R,3S,4R,5R)-5-(6-AMINO-3-METHYL-4-OXO-5H-IMIDAZO[4,5-C]PYRIDIN-1-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHOXY-HYDROXY-PHOSPHORYL] PHOSPHONO HYDROGEN PHOSPHATE | | Authors: | Chew, W.Z, Quah, S.T, Verma, C.S, Liu, Y, Lane, D.P, Brown, C.J. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Improved Eif4E Binding Peptides by Phage Display Guided Design: Plasticity of Interacting Surfaces Yield Collective Effects.
Plos One, 7, 2012
|
|
8CYI
 
 | | Cryo-EM structures and computational analysis for enhanced potency in MTA-synergic inhibition of human protein arginine methyltransferase 5 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, N-[(2-aminoquinolin-7-yl)methyl]-9-(2-hydroxyethyl)-2,3,4,9-tetrahydro-1H-carbazole-6-carboxamide, ... | | Authors: | Yadav, G.P, Wei, Z, Xiaozhi, Y, Chenglong, L, Jiang, Q. | | Deposit date: | 2022-05-23 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structure-based selection of computed ligand poses enables design of MTA-synergic PRMT5 inhibitors of better potency.
Commun Biol, 5, 2022
|
|
4U95
 
 | | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, DARPin, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pos, K.M. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB
eLife, 3, 2014
|
|
6EIF
 
 | | DYRK1A in complex with XMD7-117 | | Descriptor: | 4-(3-pyridin-3-yl-1~{H}-pyrrolo[2,3-b]pyridin-5-yl)benzenesulfonamide, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Rothweiler, U. | | Deposit date: | 2017-09-19 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Novel Scaffolds for Dual Specificity Tyrosine-Phosphorylation-Regulated Kinase (DYRK1A) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5JJA
 
 | | Crystal structure of a PP2A B56gamma/BubR1 complex | | Descriptor: | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | Authors: | Wang, Z, Wang, J, Rao, Z, Xu, W. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a PP2A B56-BubR1 complex and its implications for PP2A substrate recruitment and localization.
Protein Cell, 7, 2016
|
|
6EIJ
 
 | | DYRK1A in complex with HG-8-60-1 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ~{N}-[5-[3-(2-morpholin-4-ylethylcarbamoylamino)phenyl]-[1,3]thiazolo[5,4-b]pyridin-2-yl]cyclopropanecarboxamide | | Authors: | Rothweiler, U. | | Deposit date: | 2017-09-19 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Novel Scaffolds for Dual Specificity Tyrosine-Phosphorylation-Regulated Kinase (DYRK1A) Inhibitors.
J. Med. Chem., 61, 2018
|
|
3SV1
 
 | | Crystal structure of APP peptide bound rat Mint2 PARM | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 2, Amyloid beta A4 protein | | Authors: | Shen, Y, Long, J, Yan, X, Xie, X. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Open-closed motion of Mint2 regulates APP metabolism
J Mol Cell Biol, 5, 2013
|
|
3SUZ
 
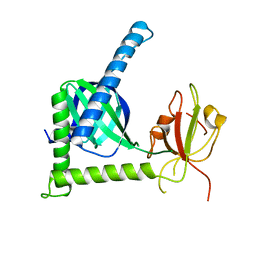 | | Crystal structure of Rat Mint2 PPC | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 2 | | Authors: | Shen, Y, Long, J, Yan, X, Xie, X. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Open-closed motion of Mint2 regulates APP metabolism
J Mol Cell Biol, 5, 2013
|
|
3NV6
 
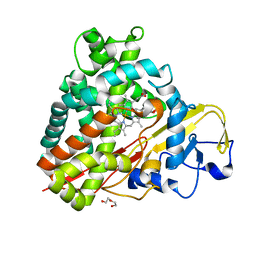 | | Crystal Structure of Camphor-Bound CYP101D2 | | Descriptor: | CAMPHOR, Cytochrome P450, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
3CGW
 
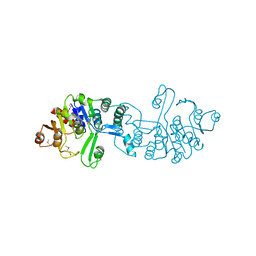 | | Crystal structure of 2-phospho-(S)-lactate transferase from Methanosarcina mazei. Northeast Structural Genomics Consortium target MaR46 | | Descriptor: | LPPG:FO 2-phospho-L-lactate transferase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Vorobiev, S.M, Ciao, M, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-06 | | Release date: | 2008-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular insights into the biosynthesis of the f420 coenzyme.
J.Biol.Chem., 283, 2008
|
|
3NV5
 
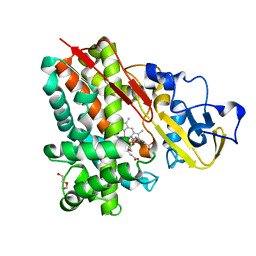 | | Crystal Structure of Cytochrome P450 CYP101D2 | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
4U8V
 
 | | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, DARPin, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pos, K.M. | | Deposit date: | 2014-08-04 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB.
Elife, 3, 2014
|
|
4U96
 
 | |
3E3R
 
 | |
1ZBP
 
 | | X-Ray Crystal Structure of Protein VPA1032 from Vibrio parahaemolyticus. Northeast Structural Genomics Consortium Target VpR44 | | Descriptor: | hypothetical protein VPA1032 | | Authors: | Forouhar, F, Yong, W, Vorobiev, S.M, Ciao, M, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
5UFI
 
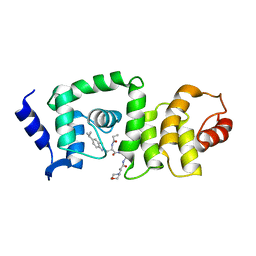 | | DCN1 bound to DI-591 | | Descriptor: | DCN1-like protein 1, N-[(1S)-1-cyclohexyl-2-{[3-(morpholin-4-yl)propanoyl]amino}ethyl]-N~2~-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alaninamide | | Authors: | Stuckey, J. | | Deposit date: | 2017-01-04 | | Release date: | 2017-11-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A potent small-molecule inhibitor of the DCN1-UBC12 interaction that selectively blocks cullin 3 neddylation.
Nat Commun, 8, 2017
|
|
4RM4
 
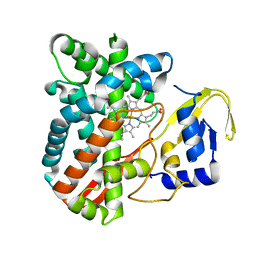 | | The crystal structure of the versatile cytochrome P450 enzyme CYP109B1 from Bacillus subtilis | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhouw, W.H, Zhang, A.L, Zhang, T, Hall, E.A, Hutchinson, S, Cryle, M.J, Wong, L.-L, Bell, S.G. | | Deposit date: | 2014-10-18 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | The crystal structure of the versatile cytochrome P450 enzyme CYP109B1 from Bacillus subtilis.
Mol Biosyst, 11, 2015
|
|
5D42
 
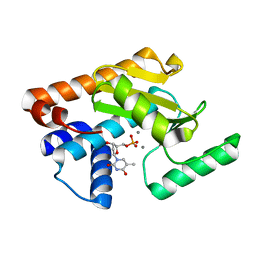 | | 2009 H1N1 PA endonuclease mutant F105S in complex with dTMP | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, THYMIDINE-5'-PHOSPHATE | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DEB
 
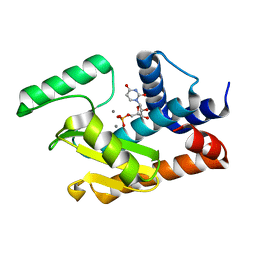 | | 2009 H1N1 PA endonuclease mutant E119D in complex with rUMP | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, URIDINE-5'-MONOPHOSPHATE | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-25 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D8U
 
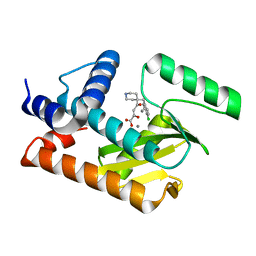 | | 2009 H1N1 PA endonuclease mutant E119D in complex with L-742,001 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-18 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D4G
 
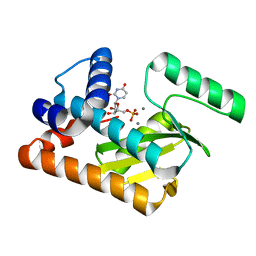 | | 2009 H1N1 PA endonuclease mutant F105S in complex with rUMP | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, URIDINE-5'-MONOPHOSPHATE | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5D9J
 
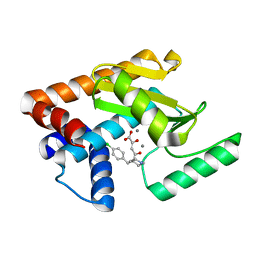 | | 2009 H1N1 PA endonuclease mutant F105S in complex with L-742,001 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-18 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2OYS
 
 | | Crystal Structure of SP1951 protein from Streptococcus pneumoniae in complex with FMN, Northeast Structural Genomics Target SpR27 | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Hypothetical protein SP1951 | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Janjua, H, Satterwhite, R, Liu, J, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional insights from structural genomics.
J.Struct.Funct.Genom., 8, 2007
|
|
2NV4
 
 | | Crystal structure of UPF0066 protein AF0241 in complex with S-adenosylmethionine. Northeast Structural Genomics Consortium target GR27 | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, UPF0066 protein AF_0241 | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Vorobiev, S.M, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-10 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional insights from structural genomics.
J.Struct.Funct.Genom., 8, 2007
|
|
