3RVG
 
 | | Crystals structure of Jak2 with a 1-amino-5H-pyrido[4,3-b]indol-4-carboxamide inhibitor | | Descriptor: | 1-(cyclohexylamino)-7-(1-methyl-1H-pyrazol-4-yl)-5H-pyrido[4,3-b]indole-4-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Lim, J, Taoka, B, Otte, R.D, Spencer, K, Dinsmore, C.J, Altman, M.D, Chan, G, Rosenstein, C, Sharma, S, Su, H.P, Szewczak, A.A, Xu, L, Yin, H, Zugay-Murphy, J, Marshall, C.G, Young, J.R. | | Deposit date: | 2011-05-06 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Discovery of 1-amino-5H-pyrido[4,3-b]indol-4-carboxamide inhibitors of Janus kinase 2 (JAK2) for the treatment of myeloproliferative disorders.
J.Med.Chem., 54, 2011
|
|
8IJY
 
 | | Synechococcus elongatus 6-4 photolyase with an 8-HDF as the antenna chromophore and a covalently linked FAD as the catalytic cofactor | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, Deoxyribodipyrimidine photolyase-related protein, ... | | Authors: | Liu, Y, Xu, L, Zhang, P. | | Deposit date: | 2023-02-28 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Synechococcus elongatus 6-4 photolyase with an 8-HDF as the antenna chromophore and a covalently linked FAD as the catalytic cofactor
To Be Published
|
|
6A96
 
 | | Cryo-EM structure of the human alpha5beta3 GABAA receptor in complex with GABA and Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-5,Gamma-aminobutyric acid receptor subunit alpha-5, ... | | Authors: | Liu, S, Xu, L, Guan, F, Liu, Y.T, Cui, Y, Zhang, Q, Bi, G.Q, Zhou, Z.H, Zhang, X, Ye, S. | | Deposit date: | 2018-07-11 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Cryo-EM structure of the human alpha 5 beta 3 GABAAreceptor.
Cell Res., 28, 2018
|
|
8IOY
 
 | | Structure of ATP7B C983S/C985S/D1027A mutant with AMP-PNP | | Descriptor: | Copper-transporting ATPase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yang, G, Xu, L, Guo, J, Wu, Z. | | Deposit date: | 2023-03-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
7V64
 
 | | Crystal structure of Antibody 16A in complex with MUC1 Glycopeptide(GlycoT) | | Descriptor: | 16A fab Heavy chain, 16A fab Light chain, 2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Niu, J, Xu, L, Meng, B, Han, Y.B, Yang, B. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Site-specific GalNAc modification on a MUC1 neoantigen epitope forms a basis for high-affinity antibody binding
To Be Published
|
|
7V7K
 
 | | Crystal structure of Antibody 16A in complex with MUC1 Glycopeptide(GlycoST) | | Descriptor: | 16A fab heavy chain, 16A fab light chain, 2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Niu, J, Xu, L, Meng, B, Han, Y.B, Yang, B. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Site-specific GalNAc modification on a MUC1 neoantigen epitope forms a basis for high-affinity antibody binding
To Be Published
|
|
7V8Q
 
 | | Crystal structure of antibody 14A in complex with MUC1 Glycopeptide(GlycoT) | | Descriptor: | 14A fab heavy chain, 14A fab light chain, 2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Niu, J, Xu, L, Meng, B, Han, Y.B, Yang, B. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Site-specific GalNAc modification on a MUC1 neoantigen epitope forms a basis for high-affinity antibody binding
To Be Published
|
|
7VAC
 
 | | Crystal structure of antibody 14A in complex with MUC1 glycopeptide(GlycoST) | | Descriptor: | 14A fab heavy chain, 14A fab light chain, 2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Niu, J, Xu, L, Meng, B, Han, Y.B, Yang, B. | | Deposit date: | 2021-08-28 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Site-specific GalNAc modification on a MUC1 neoantigen epitope forms a basis for high-affinity antibody binding
To Be Published
|
|
7VAZ
 
 | | Crystal structure of antibody 14A in complex with MUC1 glycopeptide(GlycoS) | | Descriptor: | 14A fab heavy chain, 14A fab light chain, 2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Niu, J, Xu, L, Meng, B, Han, Y.B, Yang, B. | | Deposit date: | 2021-08-30 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Site-specific GalNAc modification on a MUC1 neoantigen epitope forms a basis for high-affinity antibody binding
To Be Published
|
|
7V3Q
 
 | | Crystal structure of anti-MUC1 antibody 16A | | Descriptor: | Fab 16A Heavy Chain, Fab 16A Light Chain | | Authors: | Niu, J, Xu, L, Meng, B, Han, Y.B, Yang, B. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Site-specific GalNAc modification on a MUC1 neoantigen epitope forms a basis for high-affinity antibody binding
To Be Published
|
|
7V4W
 
 | | Crystal structure of Antibody 16A in complex with MUC1 peptide | | Descriptor: | 16A Fab Heavy chain, 16A Fab Light chain, Mucin-1 subunit alpha | | Authors: | Niu, J, Xu, L, Meng, B, Han, Y.B, Yang, B. | | Deposit date: | 2021-08-15 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Site-specific GalNAc modification on a MUC1 neoantigen epitope forms a basis for high-affinity antibody binding
To Be Published
|
|
7UIN
 
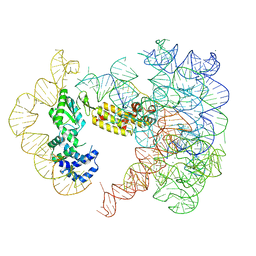 | | CryoEM Structure of an Group II Intron Retroelement | | Descriptor: | AMMONIUM ION, DNA (37-MER), E.r IIC Intron, ... | | Authors: | Chung, K, Xu, L. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of a mobile intron retroelement poised to attack its structured DNA target
Science, 378, 2022
|
|
7UIM
 
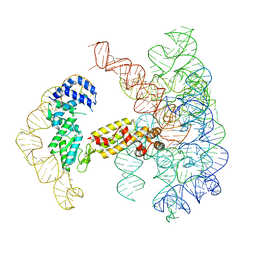 | |
7F6G
 
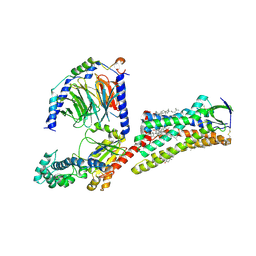 | | Cryo-EM structure of human angiotensin receptor AT1R in complex Gq proteins and Sar1-AngII | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin receptor AT1R, CHOLESTEROL, ... | | Authors: | Zhang, D, Xu, L, Zhan, Y, Guo, J, Zhang, H. | | Deposit date: | 2021-06-25 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into angiotensin receptor signaling modulation by balanced and biased agonists.
Embo J., 42, 2023
|
|
7FHN
 
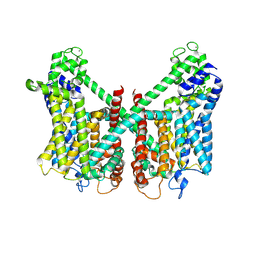 | | Structure of AtTPC1 D240A/D454A/E528A mutant with 1 mM Ca2+ | | Descriptor: | CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FHO
 
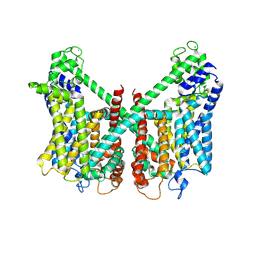 | | Structure of AtTPC1 D240A/D454A/E528A mutant with 50 mM Ca2+ | | Descriptor: | CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FHL
 
 | | Structure of AtTPC1 with 50 mM Ca2+ | | Descriptor: | AtTPC1-Cter, CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FHK
 
 | | Structure of AtTPC1 with 1 mM Ca2+ | | Descriptor: | AtTPC1-Cter, CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5XS4
 
 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5XS7
 
 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle in complex with the neutralizing antibody fragment 1D5 | | Descriptor: | Genome polyprotein, Heavy chain of Fab 1D5, Light chain of Fab 1D5 | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5XS5
 
 | | Structure of Coxsackievirus A6 (CVA6) virus procapsid particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Cheng, T, Li, S.W. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5WHZ
 
 | | PGDM1400-10E8v4 CODV Fab | | Descriptor: | Anti-HIV CODV-Fab Heavy chain, Anti-HIV CODV-Fab Light chain | | Authors: | Lord, D.M, Wei, R.R. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.549 Å) | | Cite: | Trispecific broadly neutralizing HIV antibodies mediate potent SHIV protection in macaques.
Science, 358, 2017
|
|
7DPF
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion | | Descriptor: | Capsid protein VP4, PALMITIC ACID, VP2, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7VS7
 
 | | Crystal structure of the ectodomain of OsCERK1 in complex with chitin hexamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Li, X. | | Deposit date: | 2021-10-26 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Structural insight into chitin perception by chitin elicitor receptor kinase 1 of Oryza sativa.
J Integr Plant Biol, 65, 2023
|
|
7DF6
 
 | | Mouse Galectin-3 CRD in complex with novel tetrahydropyran-based thiodisaccharide mimic inhibitor | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-5-methoxy-6-[(3R,4R,5S)-4-oxidanyl-5-(4-pyrimidin-5-yl-1,2,3-triazol-1-yl)oxan-3-yl]sulfanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-3-ol, Galectin-3 | | Authors: | Ghosh, K, Kumar, A. | | Deposit date: | 2020-11-06 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, Structure-Activity Relationships, and In Vivo Evaluation of Novel Tetrahydropyran-Based Thiodisaccharide Mimics as Galectin-3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
