2LKR
 
 | | Yeast U2/U6 complex | | Descriptor: | RNA (111-MER) | | Authors: | Burke, J.E, Sashital, D.G, Zuo, X, Wang, Y, Butcher, S.E. | | Deposit date: | 2011-10-19 | | Release date: | 2012-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the yeast U2/U6 snRNA complex.
Rna, 18, 2012
|
|
2LK3
 
 | | U2/U6 Helix I | | Descriptor: | RNA (5'-R(*GP*GP*CP*UP*UP*AP*GP*AP*UP*CP*AP*GP*AP*AP*AP*UP*GP*AP*UP*CP*AP*GP*CP*C)-3') | | Authors: | Burke, J.E, Sashital, D.G, Zuo, X.E, Wang, Y, Butcher, S.E. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the yeast U2/U6 snRNA complex.
Rna, 18, 2012
|
|
5YLS
 
 | | Crystal structure of T2R-TTL-Y50 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, E-3-(3-azanyl-4-methoxy-phenyl)-1-(5-methoxy-2,2-dimethyl-chromen-8-yl)prop-2-en-1-one, ... | | Authors: | Yang, J.H, Chen, L.J. | | Deposit date: | 2017-10-18 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 293, 2018
|
|
5YLJ
 
 | | Crystal structure of T2R-TTL-Millepachine complex | | Descriptor: | (E)-1-(5-methoxy-2,2-dimethyl-chromen-8-yl)-3-(4-methoxyphenyl)prop-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Yang, J.H, Chen, L.J. | | Deposit date: | 2017-10-17 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 293, 2018
|
|
5YWG
 
 | | Crystal structure of Arabidopsis thaliana HPPD complexed with Mesotrione | | Descriptor: | 2-[(4-methylsulfonyl-2-nitro-phenyl)-oxidanyl-methylidene]cyclohexane-1,3-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.Y, Yang, W.C. | | Deposit date: | 2017-11-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Molecular insights into the mechanism of 4-hydroxyphenylpyruvate dioxygenase inhibition: enzyme kinetics, X-ray crystallography and computational simulations.
FEBS J., 286, 2019
|
|
6ISD
 
 | | Crystal structure of Arabidopsis thaliana HPPD complexed with sulcotrione | | Descriptor: | 2-[2-chloro-4-(methylsulfonyl)benzoyl]cyclohexane-1,3-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, W.C, Yang, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into the mechanism of 4-hydroxyphenylpyruvate dioxygenase inhibition: enzyme kinetics, X-ray crystallography and computational simulations.
FEBS J., 286, 2019
|
|
5T16
 
 | |
6J63
 
 | | Crystal structure of Arabidopsis thaliana HPPD complexed with NTBC | | Descriptor: | 2-{HYDROXY[2-NITRO-4-(TRIFLUOROMETHYL)PHENYL]METHYLENE}CYCLOHEXANE-1,3-DIONE, 4-hydroxyphenylpyruvate dioxygenase, FE (III) ION | | Authors: | Yang, W.C, Yang, G.F. | | Deposit date: | 2019-01-13 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.624 Å) | | Cite: | Molecular insights into the mechanism of 4-hydroxyphenylpyruvate dioxygenase inhibition: enzyme kinetics, X-ray crystallography and computational simulations.
FEBS J., 286, 2019
|
|
3US9
 
 | | Crystal Structure of the NCX1 Intracellular Tandem Calcium Binding Domains(CBD12) | | Descriptor: | CALCIUM ION, Sodium/calcium exchanger 1 | | Authors: | Giladi, M, Sasson, Y, Hirsch, J.A, Khananshvili, D. | | Deposit date: | 2011-11-23 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A common Ca2+-driven interdomain module governs eukaryotic NCX regulation.
Plos One, 7, 2012
|
|
3WSQ
 
 | | Structure of HER2 with an Fab | | Descriptor: | Antibody Heavy Chain, Antibody Light Chain, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Fu, W.Y, Wang, Y.X, Zhou, L.J. | | Deposit date: | 2014-03-20 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insights into HER2 signaling from step-by-step optimization of anti-HER2 antibodies.
MAbs, 6, 2014
|
|
7UHE
 
 | |
6VWT
 
 | |
6VWV
 
 | |
6WJS
 
 | |
2L3R
 
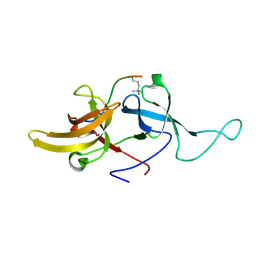 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Histone H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3 | | Authors: | Nady, N, Lemak, A, Fares, C, Gutmanas, A, Avvakumov, G, Xue, S, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-21 | | Release date: | 2011-04-13 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Recognition of Multivalent Histone States Associated with Heterochromatin by UHRF1 Protein.
J.Biol.Chem., 286, 2011
|
|
8ZBL
 
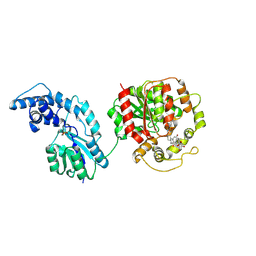 | | Glycosidated glycyrrhetinic acid derivative as a soluble epoxide hydrolase inhibitor | | Descriptor: | 1-[[(2~{S},4~{a}~{S},6~{a}~{R},6~{a}~{S},6~{b}~{R},8~{a}~{R},10~{S},12~{a}~{S},14~{b}~{S})-10-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-2,4~{a},6~{a},6~{b},9,9,12~{a}-heptamethyl-13-oxidanylidene-3,4,5,6,6~{a},7,8,8~{a},10,11,12,14~{b}-dodecahydro-1~{H}-picen-2-yl]methyl]-3-(oxan-4-ylmethyl)urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Wang, H, Feng, Y, Ge, Z.H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of glycosidated glycyrrhetinic acid derivatives: Natural product-based soluble epoxide hydrolase inhibitors.
Eur.J.Med.Chem., 280, 2024
|
|
8V1I
 
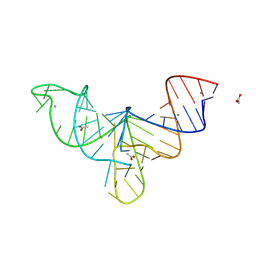 | | Crystal structure of human mascRNA | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Skeparnias, I, Zhang, J. | | Deposit date: | 2023-11-20 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of MALAT1 RNA maturation and mascRNA biogenesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V1H
 
 | | Crystal structure of human pre-mascRNA | | Descriptor: | SODIUM ION, pre-mascRNA | | Authors: | Skeparnias, I, Zhang, J. | | Deposit date: | 2023-11-20 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural basis of MALAT1 RNA maturation and mascRNA biogenesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8J17
 
 | | Crystal structure of IsPETase variant | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Yin, Q.D, Wang, Y.X. | | Deposit date: | 2023-04-12 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Efficient polyethylene terephthalate biodegradation by an engineered Ideonella sakaiensis PETase with a fixed substrate-binding W156 residue
Green Chem, 2013
|
|
6WAE
 
 | | Crystal Structure of 6X-His tagged SmcR | | Descriptor: | 1,2-ETHANEDIOL, LuxR family transcriptional regulator, SULFATE ION | | Authors: | Newman, J.D, Russell, M.M, Gonzalez-Gutierrez, G, van Kessel, J.C. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res., 49, 2021
|
|
6WAG
 
 | | Crystal Structure of SmcR S76A from Vibrio Vulnificus | | Descriptor: | 1,2-ETHANEDIOL, LuxR family transcriptional regulator, SULFATE ION | | Authors: | Newman, J.D, Russell, M.M, Gonzalez-Gutierrez, G, van Kessel, J.C. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.575 Å) | | Cite: | The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res., 49, 2021
|
|
6WAF
 
 | | Crystal Structure of SmcR N55I from Vibrio vulnificus | | Descriptor: | LuxR family transcriptional regulator, SULFATE ION | | Authors: | Newman, J.D, Russell, M.M, Gonzalez-Gutierrez, G, van Kessel, J.C. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.381 Å) | | Cite: | The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res., 49, 2021
|
|
6WAH
 
 | | Crystal Structure of SmcR L139R from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, LuxR family transcriptional regulator, SULFATE ION | | Authors: | Newman, J.D, Russell, M.M, Gonzalez-Gutierrez, G, van Kessel, J.C. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res., 49, 2021
|
|
6WAI
 
 | | Crystal Structure of SmcR N142D from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, LuxR family transcriptional regulator, SULFATE ION | | Authors: | Newman, J.D, Russell, M.M, Gonzalez-Gutierrez, G, van Kessel, J.C. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res., 49, 2021
|
|
6BOG
 
 | | Crystal structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription | | Descriptor: | RNA polymerase-associated protein RapA, SULFATE ION | | Authors: | Shaw, G.X, Gan, J, Zhou, Y.N, Zhang, R, Joachimiak, A, Jin, D.J, Ji, X. | | Deposit date: | 2017-11-20 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription.
Structure, 16, 2008
|
|
