5T4U
 
 | | Crystal structure of the bromodomain of human BRPF1 in complex with a quinolinone ligand | | Descriptor: | 1-METHYLQUINOLIN-2(1H)-ONE, NITRATE ION, Peregrin | | Authors: | Tallant, C, Igoe, N, Bayle, E.D, Nunez-Alonso, G, Newman, J.A, Mathea, S, Savitsky, P, Fedorov, O, Brennan, P.E, Muller, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fish, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-30 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of a Biased Potent Small Molecule Inhibitor of the Bromodomain and PHD Finger-Containing (BRPF) Proteins Suitable for Cellular and in Vivo Studies.
J. Med. Chem., 60, 2017
|
|
5T4V
 
 | | Crystal structure of the bromodomain of human BRPF1 in complex with NI-48 ligand | | Descriptor: | 1,2-ETHANEDIOL, 4-cyano-N-(7-methoxy-1,4-dimethyl-2-oxo-1,2-dihydroquinolin-6-yl)benzene-1-sulfonamide, FORMIC ACID, ... | | Authors: | Tallant, C, Igoe, N, Bayle, E.D, Nunez-Alonso, G, Newman, J.A, Mathea, S, Savitsky, P, Fedorov, O, Brennan, P.E, Muller, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fish, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-30 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design of a Biased Potent Small Molecule Inhibitor of the Bromodomain and PHD Finger-Containing (BRPF) Proteins Suitable for Cellular and in Vivo Studies.
J. Med. Chem., 60, 2017
|
|
4Y8D
 
 | | Crystal structure of Cyclin-G associated kinase (GAK) complexed with selective 12i inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-[3-(morpholin-4-yl)[1,2]thiazolo[4,3-b]pyridin-6-yl]aniline, Cyclin-G-associated kinase, ... | | Authors: | Chaikuad, A, Heroven, C, Nowak, R, De Jonghe, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-16 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective Inhibitors of Cyclin G Associated Kinase (GAK) as Anti-Hepatitis C Agents.
J.Med.Chem., 58, 2015
|
|
5QHV
 
 | | PanDDA analysis group deposition -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000299a | | Descriptor: | 1-cyclohexyl-3-(2-pyridin-4-ylethyl)urea, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QHY
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000462a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QHZ
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000385a | | Descriptor: | 2-cyano-~{N}-cyclohexyl-ethanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QHU
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMSOA000341b | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, N-(2-hydroxyphenyl)acetamide, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI7
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000506a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI4
 
 | | PanDDA analysis group deposition -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000466a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QI0
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000352a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QHW
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000347a | | Descriptor: | 2-methyl-~{N}-(2-methylpropyl)imidazo[1,2-a]pyridine-3-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI3
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000475a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI5
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000633a | | Descriptor: | 2-cyano-~{N}-(1,3,5-trimethylpyrazol-4-yl)ethanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI6
 
 | | PanDDA analysis group deposition -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000597a | | Descriptor: | 4-[(5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)amino]phenol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QI1
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000474a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
6Q3Z
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the inhibitor 16k | | Descriptor: | (7~{R})-2-[[2-ethoxy-4-(1-methylpiperidin-4-yl)phenyl]amino]-7-ethyl-5-methyl-8-[(4-methylthiophen-2-yl)methyl]-7~{H}-pteridin-6-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Heidenreich, D, Watts, E, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S, Hoelder, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designing Dual Inhibitors of Anaplastic Lymphoma Kinase (ALK) and Bromodomain-4 (BRD4) by Tuning Kinase Selectivity.
J.Med.Chem., 62, 2019
|
|
4XUA
 
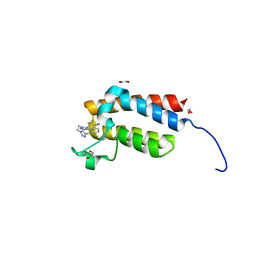 | | Crystal Structure of the bromodomain of human BAZ2B in complex with E11919 BAZ2-ICR analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-{1-[2-(4-methyl-1H-1,2,3-triazol-1-yl)ethyl]-4-phenyl-1H-imidazol-5-yl}benzonitrile, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-01-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure Enabled Design of BAZ2-ICR, A Chemical Probe Targeting the Bromodomains of BAZ2A and BAZ2B.
J.Med.Chem., 58, 2015
|
|
4XUB
 
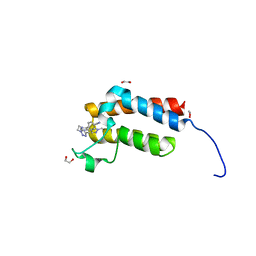 | | Crystal Structure of the bromodomain of human BAZ2B in complex with BAZ2-ICR chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-(1-methyl-1H-pyrazol-4-yl)-1-[2-(4-methyl-1H-1,2,3-triazol-1-yl)ethyl]-1H-imidazol-5-yl}benzonitrile, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-01-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure Enabled Design of BAZ2-ICR, A Chemical Probe Targeting the Bromodomains of BAZ2A and BAZ2B.
J.Med.Chem., 58, 2015
|
|
6Q3Y
 
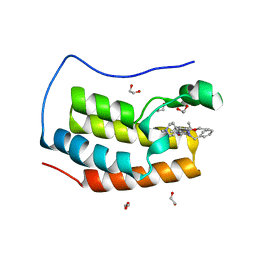 | | Crystal structure of the first bromodomain of human BRD4 in complex with the inhibitor 16i | | Descriptor: | (7~{R})-2-[[2-ethoxy-4-(1-methylpiperidin-4-yl)phenyl]amino]-7-ethyl-5-methyl-8-(phenylmethyl)-7~{H}-pteridin-6-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Heidenreich, D, Watts, E, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S, Hoelder, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Designing Dual Inhibitors of Anaplastic Lymphoma Kinase (ALK) and Bromodomain-4 (BRD4) by Tuning Kinase Selectivity.
J.Med.Chem., 62, 2019
|
|
8P08
 
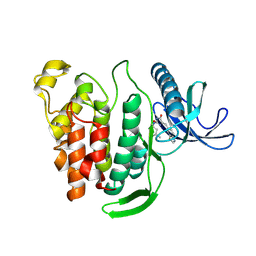 | | Crystal structure of human CLK1 in complex with Leucettinib-21 | | Descriptor: | (4~{Z})-4-(1,3-benzothiazol-6-ylmethylidene)-2-[[(2~{R})-1-methoxy-4-methyl-pentan-2-yl]amino]-1~{H}-imidazol-5-one, Dual specificity protein kinase CLK1 | | Authors: | Kraemer, A, Schroeder, M, Meijer, L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical, Biochemical, Cellular, and Physiological Characterization of Leucettinib-21, a Down Syndrome and Alzheimer's Disease Drug Candidate.
J.Med.Chem., 66, 2023
|
|
5TB6
 
 | | Structure of bromodomain of CREBBP with a pyrazolo[4,3-c]pyridin fragment | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-phenyl-1,4,6,7-tetrahydropyrazolo[4,3-c]pyridin-5-yl)propan-1-one, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Knapp, S, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
7ORF
 
 | | Crystal structure of JNK3 in complex with FMU-001-367 (compound 1) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase 10, ... | | Authors: | Chaikuad, A, Koch, P, Laufer, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-05 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Controlling the Covalent Reactivity of a Kinase Inhibitor with Light.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7OPG
 
 | | Crystal structure of CLK1 in complex with compound 2 (CC513) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-[2-(propylamino)imidazo[2,1-b][1,3,4]thiadiazol-5-yl]phenol, Dual specificity protein kinase CLK1, ... | | Authors: | Chaikuad, A, Routier, S, Bonnet, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of CLK1 in complex with compound 2 (CC513)
To Be Published
|
|
1Q3D
 
 | | GSK-3 Beta complexed with Staurosporine | | Descriptor: | GLYCOGEN SYNTHASE KINASE-3 BETA, STAUROSPORINE | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-07-29 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of the Gsk-3Beta Active Site Using Selective and Non-selective ATP-Mimetic Inhibitors
J.Mol.Biol., 333, 2003
|
|
6QAT
 
 | | Crystal structure of ULK2 in complexed with hesperadin | | Descriptor: | N-{(3Z)-2-oxo-3-[phenyl({4-[(piperidin-1-yl)methyl]phenyl}amino)methylidene]-2,3-dihydro-1H-indol-5-yl}ethanesulfonamide, Serine/threonine-protein kinase ULK2 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Biochem.J., 476, 2019
|
|
