7CNX
 
 | | Crystal structure of Apo PSD from E. coli (2.63 A) | | Descriptor: | Phosphatidylserine decarboxylase alpha chain, Phosphatidylserine decarboxylase beta chain | | Authors: | Kim, J, Cho, G. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis.
Sci Rep, 11, 2021
|
|
7CNY
 
 | | Crystal structure of 8PE bound PSD from E. coli (2.12 A) | | Descriptor: | 1,2-Dioctanoyl-SN-Glycero-3-Phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Phosphatidylserine decarboxylase alpha chain, ... | | Authors: | Kim, J, Cho, G. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis.
Sci Rep, 11, 2021
|
|
7CNZ
 
 | | Crystal structure of 10PE bound PSD from E. coli (2.70 A) | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, PHOSPHATE ION, Phosphatidylserine decarboxylase alpha chain, ... | | Authors: | Kim, J, Cho, G. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis.
Sci Rep, 11, 2021
|
|
7CNW
 
 | | Crystal structure of Apo PSD from E. coli (1.90 A) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Phosphatidylserine decarboxylase alpha chain, Phosphatidylserine decarboxylase beta chain, ... | | Authors: | Kim, J, Cho, G. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis.
Sci Rep, 11, 2021
|
|
5ZW3
 
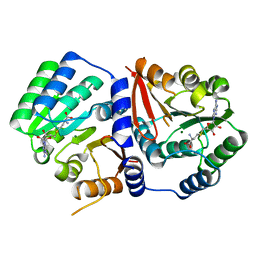 | | Crystal Structure of TrmR from B. subtilis | | Descriptor: | MAGNESIUM ION, Putative O-methyltransferase YrrM, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kim, J, Ryu, H, Almo, S.C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Identification of a novel tRNA wobble uridine modifying activity in the biosynthesis of 5-methoxyuridine.
Nucleic Acids Res., 46, 2018
|
|
5ZW4
 
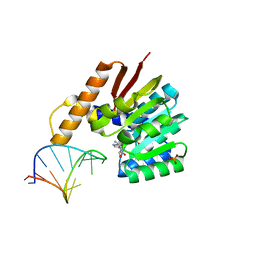 | | Crystal structure of tRNA bound TrmR | | Descriptor: | PHOSPHATE ION, Putative O-methyltransferase YrrM, RNA (5'-R(*CP*CP*UP*GP*CP*UP*UP*UP*GP*CP*AP*CP*GP*CP*AP*GP*G)-3'), ... | | Authors: | Kim, J, Ryu, H, Almo, S.C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a novel tRNA wobble uridine modifying activity in the biosynthesis of 5-methoxyuridine.
Nucleic Acids Res., 46, 2018
|
|
9IK1
 
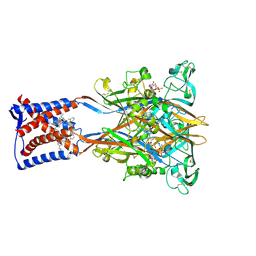 | | Cryo-EM structure of the human P2X3 receptor-compound 26a complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[2-cyclopropyl-7-[[(1~{R})-1-naphthalen-2-ylethyl]amino]-[1,2,4]triazolo[1,5-a]pyrimidin-5-yl]piperazine-1-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kim, S, Kim, G.R, Kim, Y.O, Han, X, Nagel, J, Kim, J, Song, D.I, Muller, C.E, Yoon, M.H, Jin, M.S, Kim, Y.C. | | Deposit date: | 2024-06-26 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Discovery of Triazolopyrimidine Derivatives as Selective P2X3 Receptor Antagonists Binding to an Unprecedented Allosteric Site as Evidenced by Cryo-Electron Microscopy.
J.Med.Chem., 67, 2024
|
|
8J46
 
 | | Human Consensus Olfactory Receptor OR52c in apo state, OR52c-bRIL | | Descriptor: | Olfactory receptor OR52c,Soluble cytochrome b562 | | Authors: | Choi, C.W, Bae, J, Choi, H.-J, Kim, J. | | Deposit date: | 2023-04-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
8JOZ
 
 | |
6LHW
 
 | | Structure of N-terminal and C-terminal domains of FANCA | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Kim, J, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.84 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
8ZUG
 
 | |
4AYX
 
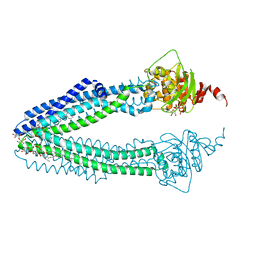 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 (ROD FORM B) | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10, CARDIOLIPIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Li, Q, Kim, J, von Delft, F, Barr, A.J, Das, S, Chaikuad, A, Xia, X, Quigley, A, Dong, Y, Dong, L, Krojer, T, Vollmar, M, Muniz, J.R.C, Bray, J.E, Berridge, G, Chalk, R, Gileadi, O, Burgess-Brown, N, Shrestha, L, Goubin, S, Yang, J, Mahajan, P, Mukhopadhyay, S, Bullock, A.N, Arrowsmith, C.H, Weigelt, J, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AYW
 
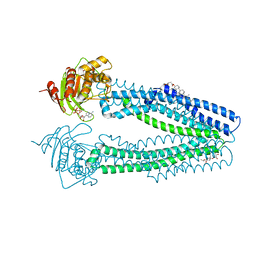 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 (PLATE FORM) | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10, CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Li, Q, Kim, J, von Delft, F, Barr, A.J, Das, S, Chaikuad, A, Xia, X, Quigley, A, Dong, Y, Dong, L, Krojer, T, Vollmar, M, Muniz, J.R.C, Bray, J.E, Berridge, G, Chalk, R, Gileadi, O, Burgess-Brown, N, Shrestha, L, Goubin, S, Yang, J, Mahajan, P, Mukhopadhyay, S, Bullock, A.N, Arrowsmith, C.H, Weigelt, J, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AYT
 
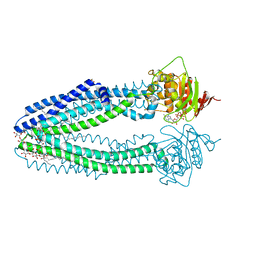 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10 MITOCHONDRIAL, CARDIOLIPIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Li, Q, Kim, J, von Delft, F, Barr, A.J, Das, S, Chaikuad, A, Xia, X, Quigley, A, Dong, Y, Dong, L, Krojer, T, Vollmar, M, Muniz, J.R.C, Bray, J.E, Berridge, G, Chalk, R, Gileadi, O, Burgess-Brown, N, Shrestha, L, Goubin, S, Yang, J, Mahajan, P, Mukhopadhyay, S, Bullock, A.N, Arrowsmith, C.H, Weigelt, J, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8TOO
 
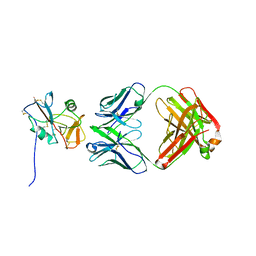 | | Crystal structure of Epstein-Barr virus gp42 in complex with antibody 4C12 | | Descriptor: | 4C12 heavy chain, 4C12 light chain, Glycoprotein 42 | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
8TNN
 
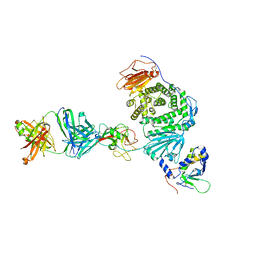 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with gp42 antibody A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A10 heavy chain, A10 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
8TNT
 
 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with antibodies F-2-1 and 769C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769C2 heavy chain, 769C2 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
4RNZ
 
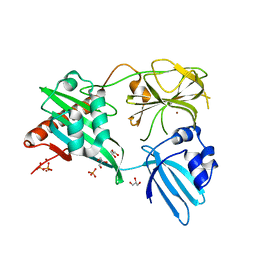 | | Structure of Helicobacter pylori Csd3 from the hexagonal crystal | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL, NICKEL (II) ION, ... | | Authors: | An, D.R, Kim, H.S, Kim, J, Im, H.N, Yoon, H.J, Yoon, J.Y, Jang, J.Y, Hesek, D, Lee, M, Mobashery, S, Kim, S.-J, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RNY
 
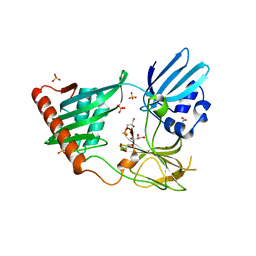 | | Structure of Helicobacter pylori Csd3 from the orthorhombic crystal | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL, SULFATE ION, ... | | Authors: | An, D.R, Kim, H.S, Kim, J, Im, H.N, Yoon, H.J, Yoon, J.Y, Jang, J.Y, Hesek, D, Lee, M, Mobashery, S, Kim, S.-J, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6LIL
 
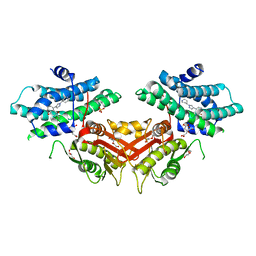 | |
8W77
 
 | | Human Consensus Olfactory Receptor OR52c in apo state, OR52c only | | Descriptor: | Human Consensus Olfactory Receptor OR52c in apo state, receptor only,Soluble cytochrome b562 | | Authors: | Choi, C.W, Bae, J, Choi, H.-J, Kim, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
3SXN
 
 | | Mycobacterium tuberculosis Eis protein initiates modulation of host immune responses by acetylation of DUSP16/MKP-7 | | Descriptor: | COENZYME A, Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J.S, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-07-15 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RYO
 
 | | Crystal Structure of Enhanced Intracellular Survival (Eis) Protein from Mycobacterium tuberculosis with Acetyl CoA | | Descriptor: | ACETYL COENZYME *A, Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-05-11 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 2012
|
|
2F0Y
 
 | |
4FYB
 
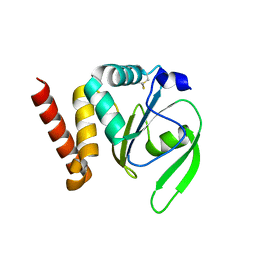 | | Structural and functional characterizations of a thioredoxin-fold protein from Helicobacter pylori | | Descriptor: | GLYCEROL, Thiol:disulfide interchange protein (DsbC) | | Authors: | Yoon, J.Y, Kim, J, Lee, S.J, Im, H.N, Kim, H.S, Yoon, H, An, D.R, Kim, J.Y, Kim, S, Han, B.W, Suh, S.W. | | Deposit date: | 2012-07-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional characterization of HP0377, a thioredoxin-fold protein from Helicobacter pylori
Acta Crystallogr.,Sect.D, 69, 2013
|
|
