4F25
 
 | |
3PT3
 
 | | Crystal structure of the C-terminal lobe of the human UBR5 HECT domain | | Descriptor: | E3 ubiquitin-protein ligase UBR5 | | Authors: | Matta-Camacho, E, Kozlov, G, Menade, M, Gehring, K. | | Deposit date: | 2010-12-02 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the HECT C-lobe of the UBR5 E3 ubiquitin ligase.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3PKN
 
 | | Crystal structure of MLLE domain of poly(A) binding protein in complex with PAM2 motif of La-related protein 4 (LARP4) | | Descriptor: | IODIDE ION, La-related protein 4, Polyadenylate-binding protein 1, ... | | Authors: | Xie, J, Kozlov, G, Gehring, K. | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | La-Related Protein 4 Binds Poly(A), Interacts with the Poly(A)-Binding Protein MLLE Domain via a Variant PAM2w Motif, and Can Promote mRNA Stability.
Mol.Cell.Biol., 31, 2011
|
|
4ZYN
 
 | | Crystal Structure of Parkin E3 ubiquitin ligase (linker deletion; delta 86-130) | | Descriptor: | E3 ubiquitin-protein ligase parkin, SULFATE ION, ZINC ION | | Authors: | Lilov, A, Sauve, V, Trempe, J.F, Rodionov, D, Wang, J, Gehring, K. | | Deposit date: | 2015-05-21 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | A Ubl/ubiquitin switch in the activation of Parkin.
Embo J., 34, 2015
|
|
3PDZ
 
 | |
4GWR
 
 | |
4Z2Z
 
 | |
3RG0
 
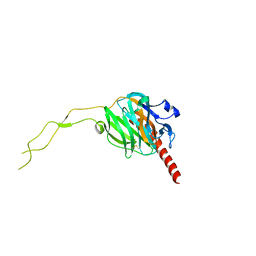 | | Structural and functional relationships between the lectin and arm domains of calreticulin | | Descriptor: | CALCIUM ION, Calreticulin | | Authors: | Kozlov, G, Pocanschi, C.L, Brockmeier, U, Williams, D.B, Gehring, K. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural and Functional Relationships between the Lectin and Arm Domains of Calreticulin.
J.Biol.Chem., 286, 2011
|
|
4EF0
 
 | |
4F26
 
 | |
4I6X
 
 | |
3KTR
 
 | |
3KTP
 
 | | Structural basis of GW182 recognition by poly(A)-binding protein | | Descriptor: | Polyadenylate-binding protein 1, Trinucleotide repeat-containing gene 6C protein | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2009-11-25 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the Mlle domain of poly(A)-binding protein.
J.Biol.Chem., 285, 2010
|
|
3KUT
 
 | |
3KUJ
 
 | |
3KUS
 
 | |
3KUI
 
 | |
3KUR
 
 | |
3NTW
 
 | |
3NY1
 
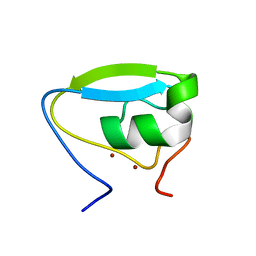 | | Structure of the ubr-box of the UBR1 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3O0X
 
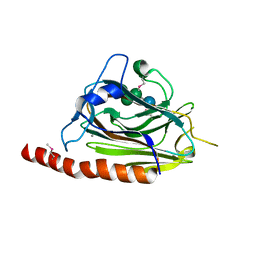 | | Structural basis of carbohydrate recognition by calreticulin | | Descriptor: | CALCIUM ION, Calreticulin, alpha-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of carbohydrate recognition by calreticulin.
J.Biol.Chem., 285, 2010
|
|
3NY2
 
 | | Structure of the ubr-box of UBR2 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR2, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3O0W
 
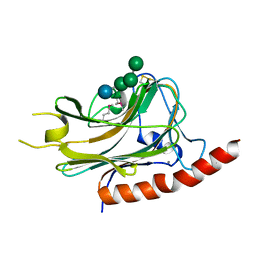 | | Structural basis of carbohydrate recognition by calreticulin | | Descriptor: | CALCIUM ION, Calreticulin, alpha-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of carbohydrate recognition by calreticulin.
J.Biol.Chem., 285, 2010
|
|
3NY3
 
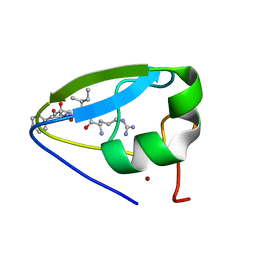 | | Structure of the ubr-box of UBR2 in complex with N-degron | | Descriptor: | E3 ubiquitin-protein ligase UBR2, N-degron, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1DU6
 
 | |
