7O33
 
 | | Crystal structure of the anti-PAS Fab 3.1 in complex with its epitope peptide | | Descriptor: | APSA epitope peptide, anti-PAS Fab 3.1 chimeric heavy chain, anti-PAS Fab 3.1 chimeric light chain | | Authors: | Schilz, J, Skerra, A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of structurally disordered Pro/Ala-rich sequences (PAS) by antibodies involves an Ala residue at the hot spot of the epitope.
J.Mol.Biol., 433, 2021
|
|
3FC2
 
 | | PLK1 in complex with BI6727 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bader, G. | | Deposit date: | 2008-11-21 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | BI 6727, A Polo-like Kinase Inhibitor with Improved Pharmacokinetic Profile and Broad Antitumor Activity.
Clin.Cancer Res., 15, 2009
|
|
1LDT
 
 | | COMPLEX OF LEECH-DERIVED TRYPTASE INHIBITOR WITH PORCINE TRYPSIN | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPTASE INHIBITOR | | Authors: | Stubbs, M.T. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The three-dimensional structure of recombinant leech-derived tryptase inhibitor in complex with trypsin. Implications for the structure of human mast cell tryptase and its inhibition.
J.Biol.Chem., 272, 1997
|
|
6Q04
 
 | | MERS-CoV S structure in complex with 5-N-acetyl neuraminic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Q06
 
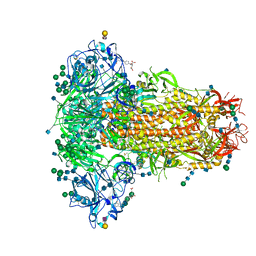 | | MERS-CoV S structure in complex with 2,3-sialyl-N-acetyl-lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Q05
 
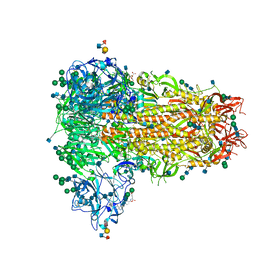 | | MERS-CoV S structure in complex with sialyl-lewisX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Q07
 
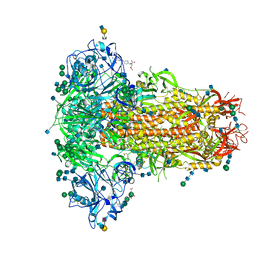 | | MERS-CoV S structure in complex with 2,6-sialyl-N-acetyl-lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7ALQ
 
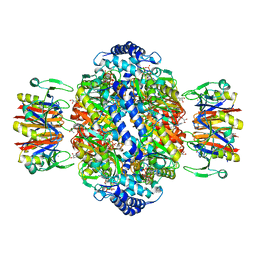 | | human GCH-GFRP inhibitory complex 7-deaza-GTP bound | | Descriptor: | 7,8-DIHYDROBIOPTERIN, 7-deaza-GTP, GTP cyclohydrolase 1, ... | | Authors: | Ebenhoch, R, Nar, H. | | Deposit date: | 2020-10-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Biophysical and structural investigation of the regulation of human GTP cyclohydrolase I by its regulatory protein GFRP.
J.Struct.Biol., 213, 2021
|
|
2ODR
 
 | | Methanococcus Maripaludis Phosphoseryl-tRNA synthetase | | Descriptor: | phosphoseryl-tRNA synthetase | | Authors: | Steitz, T.A, Kamtekar, S. | | Deposit date: | 2006-12-26 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.228 Å) | | Cite: | Toward understanding phosphoseryl-tRNACys formation: the crystal structure of Methanococcus maripaludis phosphoseryl-tRNA synthetase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1AF0
 
 | | SERRATIA PROTEASE IN COMPLEX WITH INHIBITOR | | Descriptor: | CALCIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-hydroxy-L-alaninamide, SERRATIA PROTEASE, ... | | Authors: | Baumann, U. | | Deposit date: | 1997-03-20 | | Release date: | 1998-03-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the 50 kDa Metallo Protease from S. Marcescens in Complex with the Synthetic Inhibitor Cbz-Leu-Ala-Nhoh
To be Published
|
|
5G4N
 
 | | Crystal structure of the p53 cancer mutant Y220C in complex with a difluorinated derivative of the small molecule stabilizer Phikan083 | | Descriptor: | 1-[9-(2,2-difluoroethyl)-9H-carbazol-3-yl]-N-methylmethanamine, CELLULAR TUMOR ANTIGEN P53, GLYCEROL, ... | | Authors: | Joerger, A.C, Bauer, M, Jones, R.N, Spencer, J. | | Deposit date: | 2016-05-13 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Harnessing Fluorine-Sulfur Contacts and Multipolar Interactions for the Design of P53 Mutant Y220C Rescue Drugs.
Acs Chem.Biol., 11, 2016
|
|
5G4O
 
 | | Crystal structure of the p53 cancer mutant Y220C in complex with a trifluorinated derivative of the small molecule stabilizer Phikan083 | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, N,N-dimethyl-1-[9-(2,2,2-trifluoroethyl)-9H-carbazol-3-yl]methanamine, ZINC ION | | Authors: | Joerger, A.C, Bauer, M, Baud, M.G.J, Spencer, J. | | Deposit date: | 2016-05-13 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Harnessing Fluorine-Sulfur Contacts and Multipolar Interactions for the Design of P53 Mutant Y220C Rescue Drugs.
Acs Chem.Biol., 11, 2016
|
|
5G4M
 
 | |
4E7R
 
 | | Thrombin in complex with 3-amidinophenylalanine inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(2S)-3-[4-(2-aminoethyl)piperidin-1-yl]-2-{[(2',4'-dichlorobiphenyl-3-yl)sulfonyl]amino}-3-oxopropyl]benzenecarboximidamide, GLYCEROL, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2012-03-19 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | New 3-amidinophenylalanine-derived inhibitors of matriptase
MEDCHEMCOMM, 3, 2012
|
|
1TBQ
 
 | |
1TBR
 
 | |
5LAW
 
 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND 14 | | Descriptor: | 2-[(3~{S},3'~{a}~{S},6'~{S},6'~{a}~{S})-6-chloranyl-6'-(3-chlorophenyl)-4'-(cyclopropylmethyl)-2-oxidanylidene-spiro[1~{H}-indole-3,5'-3,3~{a},6,6~{a}-tetrahydro-2~{H}-pyrrolo[3,2-b]pyrrole]-1'-yl]ethanoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
5LAZ
 
 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND BI-0252 | | Descriptor: | 4-[(2~{R},3~{a}~{S},5~{S},6~{S},6~{a}~{S})-6'-chloranyl-6-(3-chloranyl-2-fluoranyl-phenyl)-4-(cyclopropylmethyl)-2'-oxidanylidene-spiro[1,2,3,3~{a},6,6~{a}-hexahydropyrrolo[3,2-b]pyrrole-5,3'-1~{H}-indole]-2-yl]benzoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION, ... | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
5LAV
 
 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) in complex with compound 6b | | Descriptor: | (3~{S},3'~{S},4'~{S})-4'-azanyl-6-chloranyl-3'-(3-chlorophenyl)-1'-(2,2-dimethylpropyl)spiro[1~{H}-indole-3,2'-pyrrolidine]-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
5LAY
 
 | | Discovery of New Natural-product-inspired Spiro-oxindole Compounds as Orally Active Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND 6g | | Descriptor: | (3~{S},3'~{S},4'~{S},5'~{S})-4'-azanyl-6-chloranyl-3'-(3-chloranyl-2-fluoranyl-phenyl)-1'-[(3-ethoxyphenyl)methyl]-5'-methyl-spiro[1~{H}-indole-3,2'-pyrrolidine]-2-one, E3 ubiquitin-protein ligase Mdm2, GLYCEROL, ... | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
6YJX
 
 | | Structure of Hen egg-white lysozyme crystallized with PAS polypeptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Proline/alanine-rich sequence (PAS) polypeptides as an alternative to PEG precipitants for protein crystallization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6YJW
 
 | | Structure of Fragaria ananassa O-methyltransferase crystallized with PAS polypeptide | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, O-methyltransferase, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Proline/alanine-rich sequence (PAS) polypeptides as an alternative to PEG precipitants for protein crystallization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
1TFX
 
 | |
5N47
 
 | |
7JJC
 
 | |
