7MUA
 
 | | Structure of the adeno-associated virus 9 capsid at pH pH 5.5 in complex with terminal galactose | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
3OAH
 
 | |
7MTG
 
 | | Structure of the adeno-associated virus 9 capsid at pH 6.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTP
 
 | | Structure of the adeno-associated virus 9 capsid at pH 5.5 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTW
 
 | | Structure of the adeno-associated virus 9 capsid at pH 4.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
8TXO
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound to the unnatural dZ-PTP base pair in the active site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Aiyer, S, Shan, Z, Oh, J, Wang, D, Tan, Y.Z, Lyumkis, D. | | Deposit date: | 2023-08-23 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Overcoming resolution attenuation during tilted cryo-EM data collection.
Nat Commun, 15, 2024
|
|
7RL1
 
 | | AAVrh.10-7x capsid | | Descriptor: | Capsid protein VP1, DNA (5'-D(*CP*A)-3') | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural Study of Aavrh.10 Receptor and Antibody Interactions.
J.Virol., 95, 2021
|
|
7S1W
 
 | | The AAVrh.10-glycan complex | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2021-09-02 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural Study of Aavrh.10 Receptor and Antibody Interactions.
J.Virol., 95, 2021
|
|
4XZ5
 
 | | Structure of the thermostable alpha-Carbonic Anydrase from Thiomicrospira crunogena XCL-2 gammaproteobacterium | | Descriptor: | BICARBONATE ION, Carbonic anhydrase, alpha family, ... | | Authors: | Mahon, B.P, Diaz-Torres, N.A, Pinard, M.A, McKenna, R. | | Deposit date: | 2015-02-03 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structural and biophysical characterization of the alpha-carbonic anhydrase from the gammaproteobacterium Thiomicrospira crunogena XCL-2: insights into engineering thermostable enzymes for CO2 sequestration.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2QA0
 
 | | Structure of Adeno-Associated virus serotype 8 | | Descriptor: | Capsid protein, SODIUM ION | | Authors: | Nam, H.-J. | | Deposit date: | 2007-06-14 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of adeno-associated virus serotype 8, a gene therapy vector.
J.Virol., 81, 2007
|
|
7U97
 
 | | SAAV pH 4.0 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
7U95
 
 | | SAAV pH 6.0 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
7U96
 
 | | SAAV pH 5.5 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
7U94
 
 | | SAAV pH 7.4 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
3EKI
 
 | | Structural insights of the Mycoplasma hyorhinis protein Mh-p37: A putative thiamine pyrophosphate transporter | | Descriptor: | BROMIDE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Sippel, K.H, Robbins, A.H, Reutzel, R, McKenna, R. | | Deposit date: | 2008-09-19 | | Release date: | 2009-06-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the extracytoplasmic thiamine-binding lipoprotein p37 of Mycoplasma hyorhinis
J.Bacteriol., 191, 2009
|
|
5DVX
 
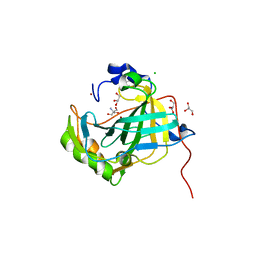 | | Crystal structure of the catalytic-domain of human carbonic anhydrase IX at 1.6 angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Carbonic anhydrase 9, ... | | Authors: | Mahon, B.P, Socorro, L, Driscoll, J.M, McKenna, R. | | Deposit date: | 2015-09-21 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | The Structure of Carbonic Anhydrase IX Is Adapted for Low-pH Catalysis.
Biochemistry, 55, 2016
|
|
3IXO
 
 | |
1KVP
 
 | |
2ILI
 
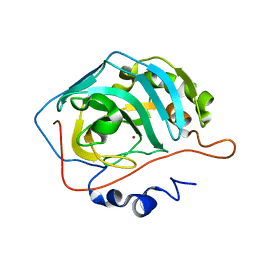 | | Refine atomic structure of human carbonic anhydrase II | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Fisher, S.Z. | | Deposit date: | 2006-10-02 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic crystal and molecular dynamics simulation structures of human carbonic anhydrase II: insights into the proton transfer mechanism
Biochemistry, 46, 2007
|
|
2AX2
 
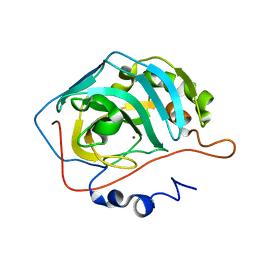 | | Production and X-ray crystallographic analysis of fully deuterated human carbonic anhydrase II | | Descriptor: | Carbonic anhydrase II, ZINC ION | | Authors: | Budayova-Spano, M, Fisher, S.Z, Dauvergne, M.T, Silverman, D.N, Myles, D.A.A, McKenna, R.M. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Production and X-ray crystallographic analysis of fully deuterated human carbonic anhydrase II.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1SGU
 
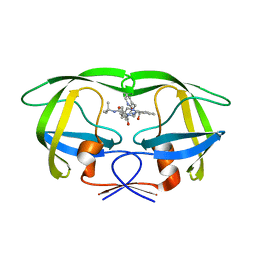 | | Comparing the Accumulation of Active Site and Non-active Site Mutations in the HIV-1 Protease | | Descriptor: | N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, POL polyprotein | | Authors: | Clemente, J.C, Moose, R.E, Hemrajani, R, Govindasamy, L, Reutzel, R, Mckenna, R, Abandje-McKenna, M, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2004-02-24 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparing the Accumulation of Active- and Nonactive-Site Mutations in the HIV-1 Protease.
Biochemistry, 43, 2004
|
|
1SH9
 
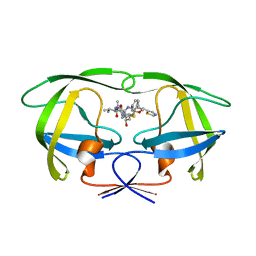 | | Comparing the Accumulation of Active Site and Non-active Site Mutations in the HIV-1 Protease | | Descriptor: | POL polyprotein, RITONAVIR | | Authors: | Clemente, J.C, Moose, R.E, Hemrajani, R, Govindasamy, L, Reutzel, R, McKenna, R, Abanje-McKenna, M, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2004-02-25 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparing the Accumulation of Active- and Nonactive-Site Mutations in the HIV-1 Protease.
Biochemistry, 43, 2004
|
|
1IJJ
 
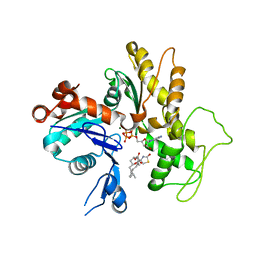 | | THE X-RAY CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN RABBIT SKELETAL MUSCLE ACTIN AND LATRUNCULIN A AT 2.85 A RESOLUTION | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Vorobiev, S.M, Bubb, M.R, Almo, S.C. | | Deposit date: | 2001-04-26 | | Release date: | 2002-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Polylysine induces an antiparallel actin dimer that nucleates filament assembly: crystal structure at 3.5-A resolution
J.Biol.Chem., 277, 2002
|
|
1G0F
 
 | |
1G0E
 
 | |
