4PBX
 
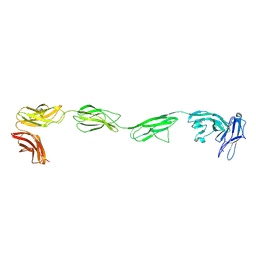 | | Crystal structure of the six N-terminal domains of human receptor protein tyrosine phosphatase sigma | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase S | | Authors: | Coles, C.H, Mitakidis, N, Zhang, P, Elegheert, J, Lu, W, Stoker, A.W, Nakagawa, T, Craig, A.M, Jones, E.Y, Aricescu, A.R. | | Deposit date: | 2014-04-14 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for extracellular cis and trans RPTP sigma signal competition in synaptogenesis.
Nat Commun, 5, 2014
|
|
4PBW
 
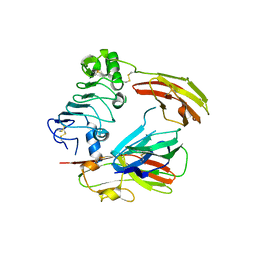 | | Crystal structure of chicken receptor protein tyrosine phosphatase sigma in complex with TrkC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NT-3 growth factor receptor, Protein-tyrosine phosphatase CRYPalpha1 isoform | | Authors: | Coles, C.H, Mitakidis, N, Zhang, P, Elegheert, J, Lu, W, Stoker, A.W, Nakagawa, T, Craig, A.M, Jones, E.Y, Aricescu, A.R. | | Deposit date: | 2014-04-14 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for extracellular cis and trans RPTP sigma signal competition in synaptogenesis.
Nat Commun, 5, 2014
|
|
4R5J
 
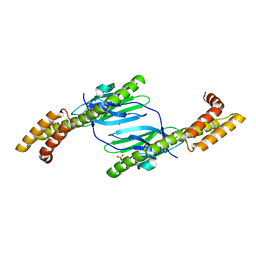 | | Crystal structure of the DnaK C-terminus (Dnak-SBD-A) | | Descriptor: | CALCIUM ION, Chaperone protein DnaK, PHOSPHATE ION | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
4R5G
 
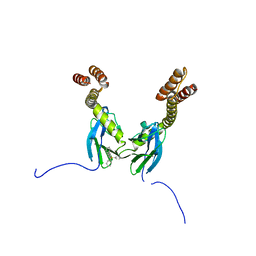 | | Crystal structure of the DnaK C-terminus with the inhibitor PET-16 | | Descriptor: | Chaperone protein DnaK, triphenyl(phenylethynyl)phosphonium | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4501 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
3P4F
 
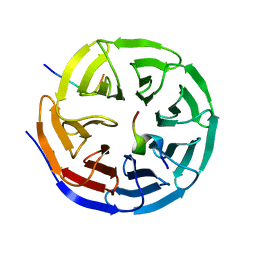 | | Structural and biochemical insights into MLL1 core complex assembly and regulation. | | Descriptor: | Histone-lysine N-methyltransferase MLL, Retinoblastoma-binding protein 5, WD repeat-containing protein 5 | | Authors: | Avdic, V, Zhang, P, Lanouette, S, Groulx, A, Tremblay, V, Brunzelle, J.B, Couture, J.-F. | | Deposit date: | 2010-10-06 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical insights into MLL1 core complex assembly.
Structure, 19, 2011
|
|
4R5L
 
 | | Crystal structure of the DnaK C-terminus (Dnak-SBD-C) | | Descriptor: | CALCIUM ION, Chaperone protein DnaK, PHOSPHATE ION, ... | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9701 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
3S32
 
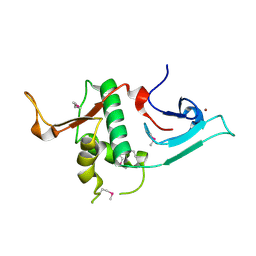 | | Crystal structure of Ash2L N-terminal domain | | Descriptor: | Set1/Ash2 histone methyltransferase complex subunit ASH2, ZINC ION | | Authors: | Sarvan, S, Avdic, V, Tremblay, V, Chaturvedi, C.-P, Zhang, P, Lanouette, S, Blais, A, Brunzelle, J.S, Brand, M, Couture, J.-F. | | Deposit date: | 2011-05-17 | | Release date: | 2011-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the trithorax group protein ASH2L reveals a forkhead-like DNA binding domain.
Nat.Struct.Mol.Biol., 18, 2011
|
|
9DCD
 
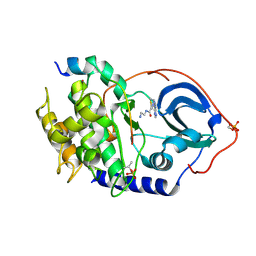 | | Structure of J-PKAc chimera in complex with Aplithianine d2 | | Descriptor: | N-(2-aminoethyl)-4-(7H-purin-6-yl)-3,4-dihydro-2H-1,4-thiazine-6-carboxamide, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Martinez Fiesco, J.A, Zhang, P. | | Deposit date: | 2024-08-25 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chemical Evolution of Aplithianine Class of Serine/Threonine Kinase Inhibitors.
J.Med.Chem., 68, 2025
|
|
9DC6
 
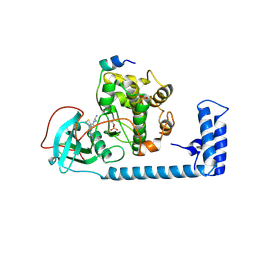 | | Structure of J-PKAc chimera in complex with Aplithianine e1 | | Descriptor: | N-(2-aminoethyl)-4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-3,4-dihydro-2H-1,4-thiazine-6-carboxamide, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Martinez Fiesco, J.A, Zhang, P. | | Deposit date: | 2024-08-25 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chemical Evolution of Aplithianine Class of Serine/Threonine Kinase Inhibitors.
J.Med.Chem., 68, 2025
|
|
6BYR
 
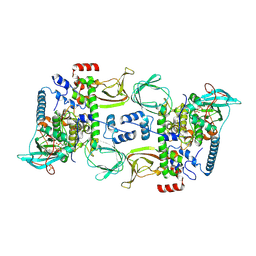 | | Structures of the PKA RI alpha holoenzyme with the FLHCC driver J-PKAc alpha or native PKAc alpha | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha chimera, MAGNESIUM ION, ... | | Authors: | Cao, B, Lu, T.W, Martinez Fiesco, J.A, Tomasini, M, Fan, L, Simon, S.M, Taylor, S.S, Zhang, P. | | Deposit date: | 2017-12-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.661 Å) | | Cite: | Structures of the PKA RI alpha Holoenzyme with the FLHCC Driver J-PKAc alpha or Wild-Type PKAc alpha.
Structure, 27, 2019
|
|
6LZ3
 
 | |
9IK4
 
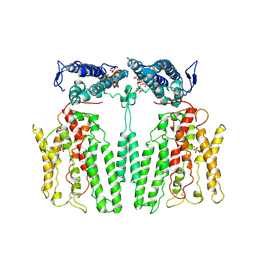 | |
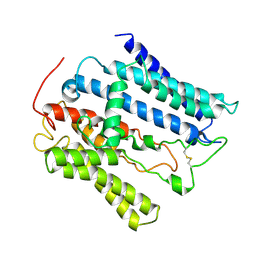
9JF8
 
 | |
6XF8
 
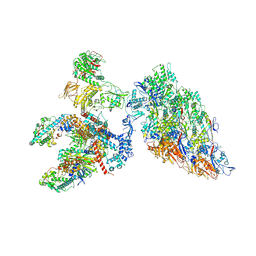 | | DLP 5 fold | | Descriptor: | Inner capsid protein lambda-1, Inner capsid protein sigma-2, Outer capsid protein mu-1, ... | | Authors: | Sutton, G, Sun, D.P, Fu, X.F, Kotecha, A, Hecksel, G.W, Clare, D.K, Zhang, P, Stuart, D, Boyce, M. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Assembly intermediates of orthoreovirus captured in the cell.
Nat Commun, 11, 2020
|
|
6SKM
 
 | | Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-13,12) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SLQ
 
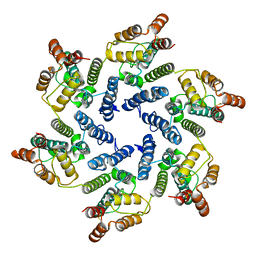 | | Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-12,11) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SLU
 
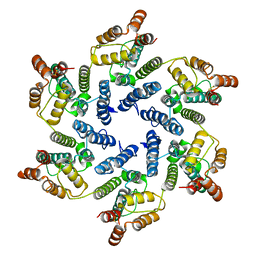 | | Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-13,11) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SMU
 
 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,12) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-22 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SKN
 
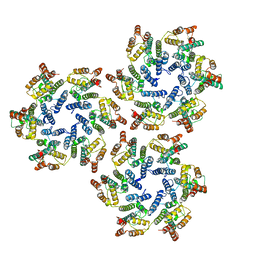 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,8) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SKK
 
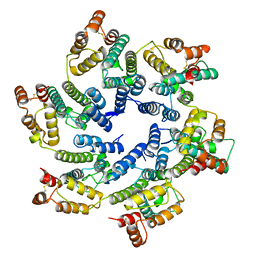 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,8) | | Descriptor: | capsid protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-15 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
9J35
 
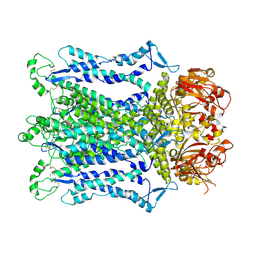 | | Cryo-EM structure of Arabidopsis CNGC5 in nanodisc | | Descriptor: | Probable cyclic nucleotide-gated ion channel 5 | | Authors: | Wang, J.P, Zhang, X, Zhang, P. | | Deposit date: | 2024-08-07 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Cryo-EM structures of Arabidopsis CNGC1 and CNGC5 reveal molecular mechanisms underlying gating and calcium selectivity.
Nat.Plants, 11, 2025
|
|
9J36
 
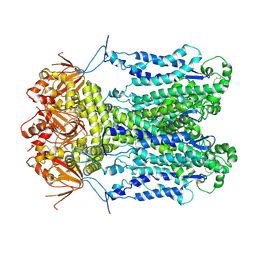 | | Cryo-EM structure of Arabidopsis CNGC5 | | Descriptor: | Probable cyclic nucleotide-gated ion channel 5 | | Authors: | Wang, J.P, Zhang, P, Zhang, X. | | Deposit date: | 2024-08-07 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structures of Arabidopsis CNGC1 and CNGC5 reveal molecular mechanisms underlying gating and calcium selectivity.
Nat.Plants, 11, 2025
|
|
9J34
 
 | | Cryo-EM structure of Arabidopsis CNGC1 | | Descriptor: | CALCIUM ION, Cyclic nucleotide-gated ion channel 1 | | Authors: | Wang, J.P, Zhang, P, Zhang, X. | | Deposit date: | 2024-08-07 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Cryo-EM structures of Arabidopsis CNGC1 and CNGC5 reveal molecular mechanisms underlying gating and calcium selectivity.
Nat.Plants, 11, 2025
|
|
9J0X
 
 | |
9J10
 
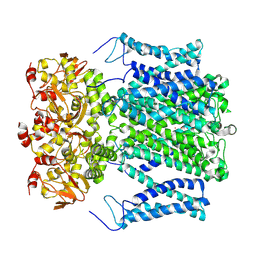 | |
