4PPA
 
 | |
4Q80
 
 | | Neutrophil serine protease 4 (PRSS57) with val-leu-lys-chloromethylketone (VLK-cmk) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-valyl-N-[(2S,3S)-7-amino-1-chloro-2-hydroxyheptan-3-yl]-L-leucinamide, ... | | Authors: | Eigenbrot, C, Lin, S.J, Dong, K.C. | | Deposit date: | 2014-04-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structures of neutrophil serine protease 4 reveal an unusual mechanism of substrate recognition by a trypsin-fold protease.
Structure, 22, 2014
|
|
4Q7Y
 
 | | Neutrophil serine protease 4 (PRSS57) apo form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Serine protease 57, ... | | Authors: | Eigenbrot, C, Lin, S.J, Dong, K.C. | | Deposit date: | 2014-04-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of neutrophil serine protease 4 reveal an unusual mechanism of substrate recognition by a trypsin-fold protease.
Structure, 22, 2014
|
|
4R69
 
 | | Lactate Dehydrogenase in complex with inhibitor compound 13 | | Descriptor: | (5R)-2-[(2-chlorophenyl)sulfanyl]-5-[2,6-dichloro-3-(tetrahydro-2H-pyran-4-ylamino)phenyl]-3-hydroxycyclohex-2-en-1-one, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Optimization of 5-(2,6-dichlorophenyl)-3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 25, 2014
|
|
4Q7X
 
 | | Neutrophil serine protease 4 (PRSS57) apo form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Serine protease 57, ... | | Authors: | Eigenbrot, C, Lin, S.J, Dong, K.C. | | Deposit date: | 2014-04-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of neutrophil serine protease 4 reveal an unusual mechanism of substrate recognition by a trypsin-fold protease.
Structure, 22, 2014
|
|
4R68
 
 | | Lactate Dehydrogenase in complex with inhibitor compound 31 | | Descriptor: | (1S)-1-phenylethyl (4-chloro-3-{[(4S)-4-(2,6-dichlorophenyl)-2-hydroxy-6-oxocyclohex-1-en-1-yl]sulfanyl}phenyl)acetate, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Optimization of 5-(2,6-dichlorophenyl)-3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 25, 2014
|
|
4PP9
 
 | |
4RLS
 
 | | Lactate Dehydrogenase in complex with inhibitor compound 47 | | Descriptor: | (1R)-5'-[(2-chlorophenyl)sulfanyl]-4'-hydroxy-2,3-dihydrospiro[indene-1,2'-pyran]-6'(3'H)-one, (2S)-2-HYDROXYPROPANOIC ACID, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Eigenbrot, C, Ultsch, M.H. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Identification of 3,6-disubstituted dihydropyrones as inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4Q7Z
 
 | | Neutrophil serine protease 4 (PRSS57) with phe-phe-arg-chloromethylketone (FFR-cmk) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Eigenbrot, C, Lin, S.J, Dong, K.C. | | Deposit date: | 2014-04-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of neutrophil serine protease 4 reveal an unusual mechanism of substrate recognition by a trypsin-fold protease.
Structure, 22, 2014
|
|
3K2U
 
 | | Crystal structure of HGFA in complex with the allosteric inhibitory antibody Fab40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody, Fab fragment, ... | | Authors: | Ganesan, R, Eigenbrot, C, Shia, S. | | Deposit date: | 2009-09-30 | | Release date: | 2009-12-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Unraveling the allosteric mechanism of serine protease inhibition by an antibody.
Structure, 17, 2009
|
|
6XE4
 
 | | BTK Fluorocyclopropyl amide inhibitor, Compound 25 | | Descriptor: | (1S,2S)-N-[2'-(6-tert-butyl-8-fluoro-1-oxophthalazin-2(1H)-yl)-3'-(hydroxymethyl)-1-methyl-6-oxo[1,6-dihydro[3,4'-bipyridine]]-5-yl]-2-fluorocyclopropane-1-carboxamide, SULFATE ION, Tyrosine-protein kinase BTK | | Authors: | Kiefer, J.R, Crawford, J.J, Lee, W, Eigenbrot, C, Yu, C. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Stereochemical Differences in Fluorocyclopropyl Amides Enable Tuning of Btk Inhibition and Off-Target Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
6XLO
 
 | | Crystal structure of bRaf in complex with inhibitor | | Descriptor: | 3-(2-cyanopropan-2-yl)-N-[2-fluoro-4-methyl-5-(7-methyl-8-oxo-7,8-dihydropyrido[2,3-d]pyridazin-3-yl)phenyl]benzamide, IODIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Yin, J, Eigenbrot, C, Wang, W. | | Deposit date: | 2020-06-28 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Targeting KRAS Mutant Cancers via Combination Treatment: Discovery of a 5-Fluoro-4-(3 H )-quinazolinone Aryl Urea pan-RAF Kinase Inhibitor.
J.Med.Chem., 64, 2021
|
|
8EXL
 
 | | Crystal structure of PI3K-alpha in complex with taselisib | | Descriptor: | 2-methyl-2-(4-{2-[3-methyl-1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-1H-pyrazol-1-yl)propanamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXO
 
 | | Crystal structure of PI3K-alpha in complex with compound 19 | | Descriptor: | 1-{(4S,11aM)-2-[(4R)-2-oxo-4-(propan-2-yl)-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-prolinamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXU
 
 | | Crystal structure of PI3K-alpha in complex with compound 30 | | Descriptor: | (2S)-2-cyclopropyl-2-({(4S,11aM)-2-[(4S)-2-oxo-4-(trifluoromethyl)-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}amino)acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXV
 
 | | Crystal structure of PI3K-alpha in complex with compound 32 | | Descriptor: | N~2~-{(4S,11aP)-2-[(4S)-4-(difluoromethyl)-2-oxo-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-alaninamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
1YGC
 
 | | Short Factor VIIa with a small molecule inhibitor | | Descriptor: | (R)-4-[2-(3-AMINO-BENZENESULFONYLAMINO)-1-(3,5-DIETHOXY-2-FLUOROPHENYL)-2-OXO-ETHYLAMINO]-2-HYDROXY-BENZAMIDINE, CALCIUM ION, Coagulation factor VII, ... | | Authors: | Olivero, A.G, Eigenbrot, C, Goldsmith, R, Robarge, K, Artis, D.R, Flygare, J, Rawson, T, Refino, C, Bunting, S, Kirchhofer, D. | | Deposit date: | 2005-01-04 | | Release date: | 2005-01-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A selective, slow binding inhibitor of factor VIIa binds to a nonstandard active site conformation and attenuates thrombus formation in vivo.
J.Biol.Chem., 280, 2005
|
|
2WUC
 
 | | Crystal structure of HGFA in complex with the allosteric non- inhibitory antibody Fab40.deltaTrp and Ac-KQLR-chloromethylketone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACE-KQLR-CHLOROMETHYLKETONE INHIBITOR, FAB FRAGMENT FAB40.DELTATRP HEAVY CHAIN, ... | | Authors: | Ganesan, R, Eigenbrot, C, Shia, S. | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unraveling the Allosteric Mechanism of Serine Protease Inhibition by an Antibody
Structure, 17, 2009
|
|
2WUB
 
 | | Crystal structure of HGFA in complex with the allosteric non- inhibitory antibody Fab40.deltaTrp | | Descriptor: | FAB FRAGMENT FAB40.DELTATRP HEAVY CHAIN, FAB FRAGMENT FAB40.DELTATRP LIGHT CHAIN, HEPATOCYTE GROWTH FACTOR ACTIVATOR LONG CHAIN, ... | | Authors: | Ganesan, R, Eigenbrot, C, Shia, S. | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Unraveling the Allosteric Mechanism of Serine Protease Inhibition by an Antibody
Structure, 17, 2009
|
|
1QE6
 
 | | INTERLEUKIN-8 WITH AN ADDED DISULFIDE BETWEEN RESIDUES 5 AND 33 (L5C/H33C) | | Descriptor: | INTERLEUKIN-8 VARIANT, SULFATE ION | | Authors: | Gerber, N, Lowman, H, Artis, D.R, Eigenbrot, C. | | Deposit date: | 1999-07-13 | | Release date: | 2000-03-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Receptor-binding conformation of the "ELR" motif of IL-8: X-ray structure of the L5C/H33C variant at 2.35 A resolution.
Proteins, 38, 2000
|
|
1M14
 
 | |
1M17
 
 | |
1YBW
 
 | | Protease domain of HGFA with no inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hepatocyte growth factor activator precursor | | Authors: | Shia, S, Stamos, J, Kirchhofer, D, Fan, B, Wu, J, Corpuz, R.T, Santell, L, Lazarus, R.A, Eigenbrot, C. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational lability in serine protease active sites: structures of hepatocyte growth factor activator (HGFA) alone and with the inhibitory domain from HGFA inhibitor-1B.
J.Mol.Biol., 346, 2005
|
|
1YC0
 
 | | short form HGFA with first Kunitz domain from HAI-1 | | Descriptor: | Hepatocyte growth factor activator, Kunitz-type protease inhibitor 1, PHOSPHATE ION | | Authors: | Shia, S, Stamos, J, Kirchhofer, D, Fan, B, Wu, J, Corpuz, R.T, Santell, L, Lazarus, R.A, Eigenbrot, C. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational lability in serine protease active sites: structures of hepatocyte growth factor activator (HGFA) alone and with the inhibitory domain from HGFA inhibitor-1B.
J.Mol.Biol., 346, 2005
|
|
1AAP
 
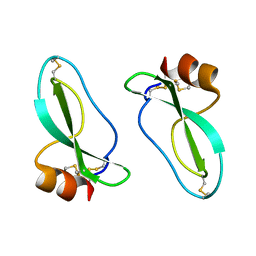 | | X-RAY CRYSTAL STRUCTURE OF THE PROTEASE INHIBITOR DOMAIN OF ALZHEIMER'S AMYLOID BETA-PROTEIN PRECURSOR | | Descriptor: | ALZHEIMER'S DISEASE AMYLOID A4 PROTEIN | | Authors: | Hynes, T.R, Randal, M, Kennedy, L.A, Eigenbrot, C, Kossiakoff, A.A. | | Deposit date: | 1990-09-14 | | Release date: | 1991-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of the protease inhibitor domain of Alzheimer's amyloid beta-protein precursor.
Biochemistry, 29, 1990
|
|
