2WCB
 
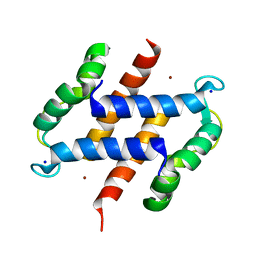 | | S100A12 complex with zinc in the absence of calcium | | Descriptor: | PROTEIN S100-A12, SODIUM ION, ZINC ION | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
2WCF
 
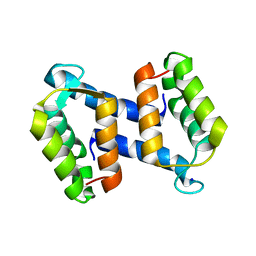 | | calcium-free (apo) S100A12 | | Descriptor: | PROTEIN S100-A12, SODIUM ION | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
3FQ9
 
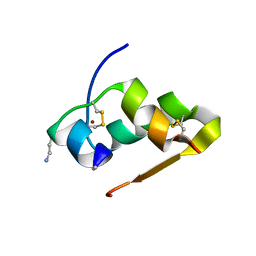 | | Design of an insulin analog with enhanced receptor-binding selectivity. Rationale, structure, and therapeutic implications | | Descriptor: | Insulin, ZINC ION | | Authors: | Zhao, M, Wan, Z.L, Whittaker, L, Xu, B, Phillips, N, Katsoyannis, P, Whittaker, J, Weiss, M.A. | | Deposit date: | 2009-01-07 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design of an insulin analog with enhanced receptor binding selectivity: rationale, structure, and therapeutic implications.
J.Biol.Chem., 284, 2009
|
|
2WC8
 
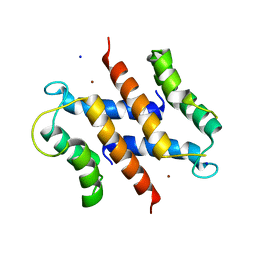 | | S100A12 complex with zinc in the absence of calcium | | Descriptor: | CITRIC ACID, PROTEIN S100-A12, SODIUM ION, ... | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
1MYI
 
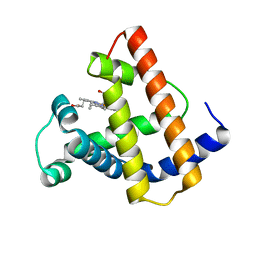 | | HIGH RESOLUTION X-RAY STRUCTURES OF PIG METMYOGLOBIN AND TWO CD3 MUTANTS MB(LYS45-> ARG) AND MB(LYS45-> SER) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Smerdon, S.J, Oldfield, T.J, Wilkinson, A.J, Dauter, Z, Petratos, K, Wilson, K.S. | | Deposit date: | 1992-02-27 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution X-ray structures of pig metmyoglobin and two CD3 mutants: Mb(Lys45----Arg) and Mb(Lys45----Ser).
Biochemistry, 31, 1992
|
|
1MYH
 
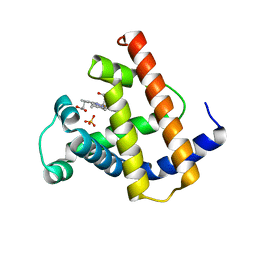 | | HIGH RESOLUTION X-RAY STRUCTURES OF PIG METMYOGLOBIN AND TWO CD3 MUTANTS MB(LYS45-> ARG) AND MB(LYS45-> SER) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Smerdon, S.J, Oldfield, T.J, Wilkinson, A.J, Dauter, Z, Petratos, K, Wilson, K.S. | | Deposit date: | 1992-02-27 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution X-ray structures of pig metmyoglobin and two CD3 mutants: Mb(Lys45----Arg) and Mb(Lys45----Ser).
Biochemistry, 31, 1992
|
|
1XW7
 
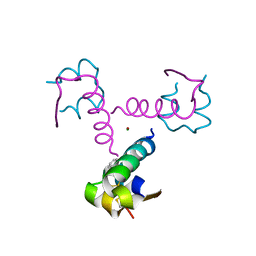 | | Diabetes-Associated Mutations in Human Insulin: Crystal Structure and Photo-Cross-Linking Studies of A-Chain Variant Insulin Wakayama | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z.L, Huang, K, Xu, B, Chu, Y.C, Hu, S.Q, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2004-10-29 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Diabetes-associated mutations in human insulin: crystal structure and photo-cross-linking studies of a-chain variant insulin wakayama
Biochemistry, 44, 2005
|
|
1WAP
 
 | |
3BXQ
 
 | | The structure of a mutant insulin uncouples receptor binding from protein allostery. An electrostatic block to the TR transition | | Descriptor: | ZINC ION, insulin A chain, insulin B chain | | Authors: | Wan, Z.L, Huang, K, Hu, S.Q, Whittaker, J, Weiss, M.A. | | Deposit date: | 2008-01-14 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of a mutant insulin uncouples receptor binding from protein allostery. An electrostatic block to the TR transition.
J.Biol.Chem., 283, 2008
|
|
3KQ6
 
 | | Enhancing the Therapeutic Properties of a Protein by a Designed Zinc-Binding Site, Structural principles of a novel long-acting insulin analog | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Wan, Z.L, Hu, S.Q, Whittaker, L, Phillips, N.B, Whittake, J, Ismail-Beigi, F, Weiss, M.A. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Supramolecular protein engineering: design of zinc-stapled insulin hexamers as a long acting depot.
J.Biol.Chem., 285, 2010
|
|
2HF0
 
 | | Bifidobacterium longum bile salt hydrolase | | Descriptor: | Bile salt hydrolase | | Authors: | Suresh, C.G, Kumar, R.S, Brannigan, J.A. | | Deposit date: | 2006-06-22 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Analysis of a Conjugated Bile Salt Hydrolase from Bifidobacterium longum Reveals an Evolutionary Relationship with Penicillin V Acylase.
J.Biol.Chem., 281, 2006
|
|
2HEZ
 
 | | Bifidobacterium longum bile salt hydrolase | | Descriptor: | Bile salt hydrolase, SULFATE ION | | Authors: | Suresh, C.G, Kumar, R.S, Brannigan, J.A. | | Deposit date: | 2006-06-22 | | Release date: | 2006-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Analysis of a Conjugated Bile Salt Hydrolase from Bifidobacterium longum Reveals an Evolutionary Relationship with Penicillin V Acylase.
J.Biol.Chem., 281, 2006
|
|
1TGL
 
 | | A SERINE PROTEASE TRIAD FORMS THE CATALYTIC CENTRE OF A TRIACYLGLYCEROL LIPASE | | Descriptor: | TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Brady, L, Brzozowski, A.M, Derewenda, Z.S, Dodson, E.J, Dodson, G.G, Tolley, S.P, Turkenburg, J.P, Christiansen, L, Huge-Jensen, B, Norskov, L, Thim, L. | | Deposit date: | 1990-02-05 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A serine protease triad forms the catalytic centre of a triacylglycerol lipase.
Nature, 343, 1990
|
|
2RKM
 
 | | STRUCTURE OF OPPA COMPLEXED WITH LYS-LYS | | Descriptor: | LYSINE, OLIGO-PEPTIDE BINDING PROTEIN, URANYL (VI) ION | | Authors: | Sleigh, S.H, Tame, J.R.H, Wilkinson, A.J. | | Deposit date: | 1997-03-25 | | Release date: | 1997-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide binding in OppA, the crystal structures of the periplasmic oligopeptide binding protein in the unliganded form and in complex with lysyllysine.
Biochemistry, 36, 1997
|
|
2OLB
 
 | | OLIGOPEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH TRI-LYSINE | | Descriptor: | ACETATE ION, OLIGO-PEPTIDE BINDING PROTEIN, TRIPEPTIDE LYS-LYS-LYS, ... | | Authors: | Tame, J, Wilkinson, A.J. | | Deposit date: | 1995-09-10 | | Release date: | 1996-01-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of the oligopeptide-binding protein OppA complexed with tripeptide and tetrapeptide ligands.
Structure, 3, 1995
|
|
5IQP
 
 | | 14-3-3 PROTEIN TAU ISOFORM | | Descriptor: | 14-3-3 protein theta, SULFATE ION | | Authors: | Xiao, B, Smerdon, S.J, Gamblin, S.J. | | Deposit date: | 2016-03-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways
Nature, 376, 1995
|
|
3ENG
 
 | | STRUCTURE OF ENDOGLUCANASE V CELLOBIOSE COMPLEX | | Descriptor: | ENDOGLUCANASE V CELLOBIOSE COMPLEX, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
5KQV
 
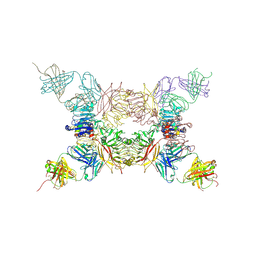 | | Insulin receptor ectodomain construct comprising domains L1,CR,L2, FnIII-1 and alphaCT peptide in complex with bovine insulin and FAB 83-14 (REVISED STRUCTURE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin, Insulin receptor,Insulin receptor, ... | | Authors: | Lawrence, M.C, Smith, B.J, Croll, T.I. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | How insulin engages its primary binding site on the insulin receptor.
Nature, 493, 2013
|
|
1OLA
 
 | |
1RKM
 
 | | STRUCTURE OF OPPA | | Descriptor: | OLIGO-PEPTIDE BINDING PROTEIN | | Authors: | Sleigh, S.H, Tame, J.R.H, Wilkinson, A.J. | | Deposit date: | 1997-03-25 | | Release date: | 1997-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Peptide binding in OppA, the crystal structures of the periplasmic oligopeptide binding protein in the unliganded form and in complex with lysyllysine.
Biochemistry, 36, 1997
|
|
4LVN
 
 | |
4LVO
 
 | |
1MNI
 
 | |
1OLC
 
 | |
1QKB
 
 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KVK | | Descriptor: | ACETATE ION, PEPTIDE LYS-VAL-LYS, PERIPLASMIC OLIGOPEPTIDE-BINDING PROTEIN, ... | | Authors: | Tame, J.R.H, Sleigh, S.H, Wilkinson, A.J. | | Deposit date: | 1999-07-14 | | Release date: | 1999-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and Calorimetric Analysis of Peptide Binding to Oppa Protein
J.Mol.Biol., 291, 1999
|
|
