1S62
 
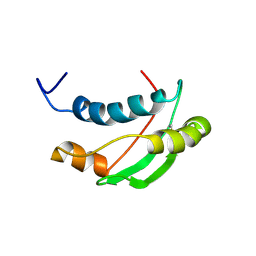 | | Solution structure of the Escherichia coli TolA C-terminal domain | | Descriptor: | TolA protein | | Authors: | Deprez, C, Blanchard, L, Simorre, J.-P, Gavioli, M, Guerlesquin, F, Lazdunski, C, Lloubes, R, Marion, D. | | Deposit date: | 2004-01-22 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the E.coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain.
J.Mol.Biol., 346, 2005
|
|
1SF5
 
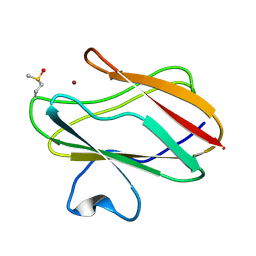 | | Structure of oxidized state of the P94A mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION | | Authors: | Carrell, C.J, Sun, D, Jiang, S, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Studies of Two Mutants of Amicyanin from Paracoccus denitrificans That Stabilize the Reduced State of the Copper.
Biochemistry, 43, 2004
|
|
1SFD
 
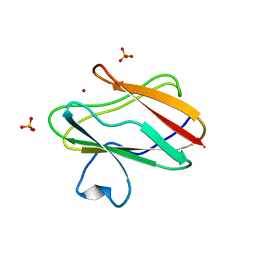 | | oxidized form of amicyanin mutant P94F | | Descriptor: | Amicyanin, COPPER (II) ION, SULFATE ION | | Authors: | Carrell, C.J, Sun, D, Jiang, S, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structural Studies of Two Mutants of Amicyanin from Paracoccus denitrificans That Stabilize the Reduced State of the Copper.
Biochemistry, 43, 2004
|
|
2SPC
 
 | | CRYSTAL STRUCTURE OF THE REPETITIVE SEGMENTS OF SPECTRIN | | Descriptor: | SPECTRIN | | Authors: | Yan, Y, Winograd, E, Viel, A, Cronin, T, Harrison, S.C, Branton, D. | | Deposit date: | 1994-03-01 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the repetitive segments of spectrin.
Science, 262, 1993
|
|
7OEK
 
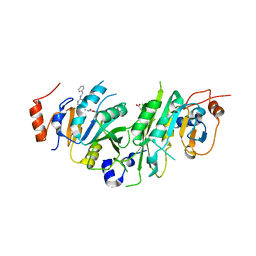 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ091 | | Descriptor: | 4-[[(3S)-3-cyclopropyl-2-azaspiro[3.3]heptan-2-yl]methyl]-2-oxidanyl-N-[[(3R)-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
1O03
 
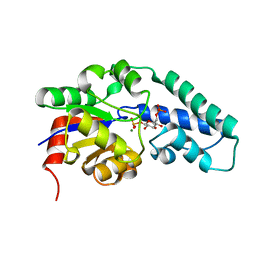 | | Structure of Pentavalent Phosphorous Intermediate of an Enzyme Catalyzed Phosphoryl transfer Reaction observed on cocrystallization with Glucose 6-phosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-02-20 | | Release date: | 2003-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The pentacovalent phosphorus intermediate of a phosphoryl transfer reaction.
Science, 299, 2003
|
|
7OEL
 
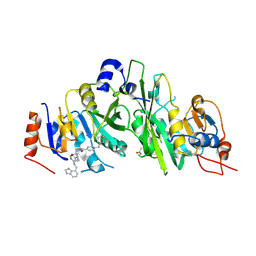 | |
7OEF
 
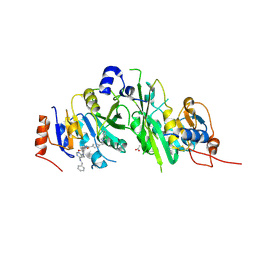 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ038 | | Descriptor: | 4-(2-azaspiro[3.4]octan-2-ylmethyl)-~{N}-[[(3~{R})-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
7OEE
 
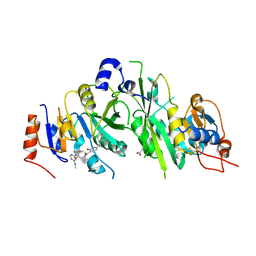 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ019b | | Descriptor: | 4-[(4,4-dimethylpiperidin-1-yl)methyl]-~{N}-[[(3~{S})-1-[6-(methylamino)pyrimidin-4-yl]-3-oxidanyl-piperidin-3-yl]methyl]-2-oxidanyl-benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
1S0P
 
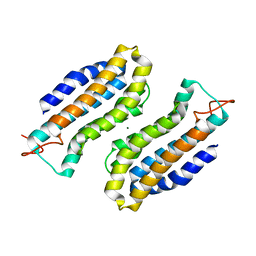 | | Structure of the N-Terminal Domain of the Adenylyl Cyclase-Associated Protein (CAP) from Dictyostelium discoideum. | | Descriptor: | Adenylyl cyclase-associated protein, MAGNESIUM ION | | Authors: | Ksiazek, D, Brandstetter, H, Israel, L, Bourenkov, G.P, Katchalova, G, Janssen, K.P, Bartunik, H.D, Noegel, A.A, Schleicher, M, Holak, T.A. | | Deposit date: | 2004-01-01 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | STRUCTURE OF THE N-TERMINAL DOMAIN OF THE ADENYLYL
CYCLASE-ASSOCIATED PROTEIN (CAP) FROM DICTYOSTELIUM DISCOIDEUM
Structure, 11, 2003
|
|
7OEI
 
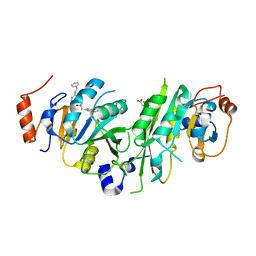 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ083 | | Descriptor: | (3~{R})-3-[[5-[(4,4-dimethylpiperidin-1-yl)methyl]-1~{H}-benzimidazol-2-yl]methyl]-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-ol, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
7OED
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ019a | | Descriptor: | 4-[(4,4-dimethylpiperidin-1-yl)methyl]-~{N}-[[(3~{R})-1-[6-(methylamino)pyrimidin-4-yl]-3-oxidanyl-piperidin-3-yl]methyl]-2-oxidanyl-benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
1S28
 
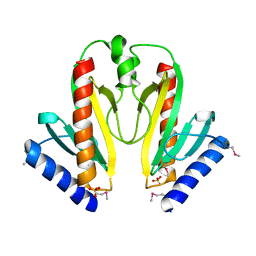 | | Crystal Structure of AvrPphF ORF1, the Chaperone for the Type III Effector AvrPphF ORF2 from P. syringae | | Descriptor: | ORF1, SULFATE ION | | Authors: | Singer, A.U, Desveaux, D, Betts, L, Chang, J.H, Nimchuk, Z, Grant, S.R, Dangl, J.L, Sondek, J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-09-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of the Type III Effector Protein AvrPphF and Its Chaperone Reveal Residues Required for Plant Pathogenesis
Structure, 12, 2004
|
|
7OEJ
 
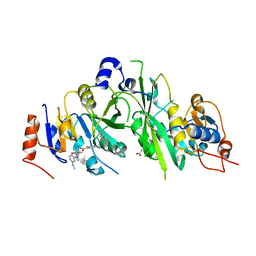 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ090 | | Descriptor: | 4-[[(3S)-3-cyclopropyl-2-azaspiro[3.3]heptan-2-yl]methyl]-N-[[(3R)-1-[6-(methylamino)pyrimidin-4-yl]-3-oxidanyl-piperidin-3-yl]methyl]-2-oxidanyl-benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
7OEH
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ059b | | Descriptor: | 4-[[(3S)-3-cyclopropyl-2-azaspiro[3.3]heptan-2-yl]methyl]-N-[[(3S)-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
7OEG
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ040b | | Descriptor: | (R)-4-((2-azaspiro[3.3]heptan-2-yl)methyl)-N-((1-(6-(benzylamino)pyrimidin-4-yl)-3-hydroxypiperidin-3-yl)methyl)benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
5KMS
 
 | |
7OEM
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ120 | | Descriptor: | 4-[(4,4-dimethylpiperidin-1-yl)methyl]-2-methylsulfanyl-~{N}-[[(3~{R})-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
2RO2
 
 | | Solution structure of domain I of the negative polarity CChMVd hammerhead ribozyme | | Descriptor: | RNA (5'-R(*GP*GP*GP*AP*GP*AP*CP*CP*UP*GP*AP*AP*GP*UP*GP*GP*GP*UP*UP*UP*CP*CP*C)-3') | | Authors: | Gallego, J, Dufour, D, Gago, S, de la Pena, M, Flores, R. | | Deposit date: | 2008-03-05 | | Release date: | 2008-12-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the ribozymes of chrysanthemum chlorotic mottle viroid: a loop-loop interaction motif conserved in most natural hammerheads
Nucleic Acids Res., 37, 2009
|
|
1ODZ
 
 | | Expansion of the glycosynthase repertoire to produce defined manno-oligosaccharides | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Mannan endo-1,4-beta-mannosidase, SODIUM ION, ... | | Authors: | Jahn, M, Stoll, D, Warren, R.A.J, Szabo, L, Singh, P, Gilbert, H.J, Ducros, V.M.A, Davies, G.J, Withers, S.G. | | Deposit date: | 2003-03-17 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expansion of the Glycosynthase Repertoire to Produce Defined Manno-Oligosaccharides
Chem.Commun.(Camb.), 12, 2003
|
|
1S0G
 
 | |
1OF0
 
 | | CRYSTAL STRUCTURE OF BACILLUS SUBTILIS COTA AFTER 1H SOAKING WITH ABTS | | Descriptor: | 3-ETHYL-2-[(2Z)-2-(3-ETHYL-6-SULFO-1,3-BENZOTHIAZOL-2(3H)-YLIDENE)HYDRAZINO]-6-SULFO-3H-1,3-BENZOTHIAZOL-1-IUM, COPPER (I) ION, CU-O LINKAGE, ... | | Authors: | Enguita, F.J, Marcal, D, Grenha, R, Martins, L.O, Henriques, A.O, Carrondo, M.A. | | Deposit date: | 2003-04-03 | | Release date: | 2004-05-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Characterization of a Bacterial Laccase Reaction Cycle
To be Published
|
|
1S96
 
 | | The 2.0 A X-ray structure of Guanylate Kinase from E.coli | | Descriptor: | Guanylate kinase, PHOSPHATE ION, UNKNOWN ATOM OR ION | | Authors: | Kreusch, A, Spraggon, G, Klock, H.E, McMullan, D, Vincent, J, Rodrigues, K, Lesley, S.A. | | Deposit date: | 2004-02-03 | | Release date: | 2004-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Guanylate Kinase from Escherichia coli at 2.0 A resolution
To be Published
|
|
5L3F
 
 | | LSD1-CoREST1 in complex with polymyxin B | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, Polmyxin B, ... | | Authors: | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | Deposit date: | 2016-04-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
1S2W
 
 | | Crystal structure of phosphoenolpyruvate mutase in high ionic strength | | Descriptor: | Phosphoenolpyruvate phosphomutase, SULFATE ION | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
