6G6M
 
 | | Crystal structure of the computationally designed Tako8 protein in P42212 | | Descriptor: | SULFATE ION, Tako8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
4CBB
 
 | | APO FORM OF BETAINE ALDEHYDE DEHYDROGENASE FROM Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Gonzalez-Segura, L, Diaz-Sanchez, A.G, Munoz-Clares, R.A. | | Deposit date: | 2013-10-11 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structural Bases of the Dual Coenzyme Specificity of Betaine Aldehyde Dehydrogenase from Pseudomonas Aeruginosa
To be Published
|
|
5YWA
 
 | |
6G0E
 
 | | BRD4 (BD1) in complex with APSC-derived ligands | | Descriptor: | 4-[2-(4-cyclohexylpiperazin-4-ium-1-yl)-2-oxidanylidene-ethyl]sulfanyl-1-ethyl-quinolin-2-one, Bromodomain-containing protein 4, FORMIC ACID, ... | | Authors: | Pretzel, J, Humbeck, L. | | Deposit date: | 2018-03-18 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.613 Å) | | Cite: | Discovery of an Unexpected Similarity in Ligand Binding Between BRD4 and PPARgamma
Chemrxiv, 2019
|
|
1NZF
 
 | | T4 phage BGT-D100A mutant in complex with UDP-glucose: Form II | | Descriptor: | CHLORIDE ION, DNA beta-glycosyltransferase, GLYCEROL, ... | | Authors: | Lariviere, L, Morera, S. | | Deposit date: | 2003-02-17 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the T4 phage beta-glucosyltransferase and the D100A mutant in complex with
UDP-glucose: glucose binding and identification of the catalytic base for a direct displacement mechanism.
J.Mol.Biol., 330, 2003
|
|
5NR7
 
 | | Mtb TMK crystal structure in complex with compound 43 | | Descriptor: | 1-[1-[[4-(3-chloranylphenoxy)quinolin-2-yl]methyl]piperidin-4-yl]-5-methyl-pyrimidine-2,4-dione, CHLORIDE ION, Thymidylate kinase | | Authors: | Merceron, R, Song, L, Munier-Lehmann, H, Van Calenbergh, S, Savvides, S. | | Deposit date: | 2017-04-22 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure Guided Lead Generation toward Nonchiral M. tuberculosis Thymidylate Kinase Inhibitors.
J. Med. Chem., 61, 2018
|
|
6G0G
 
 | |
6G6Q
 
 | | Crystal structure of the computationally designed Ika4 protein | | Descriptor: | Ika4 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
3V84
 
 | | Thaumatin by LB based Hanging Drop Vapour Diffusion after 1.81 MGy X-Ray dose at ESRF ID29 beamline (Worst Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Scudieri, D, Tripathi, S, Pechkova, E, Nicolini, C. | | Deposit date: | 2011-12-22 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-Blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
CRIT.REV.EUKARYOT.GENE EXPR., 22, 2012
|
|
8C5Y
 
 | | RPA tetrameric supercomplex from Pyrococcus abyssi | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Madru, C, Martinez-Carranza, M, Legrand, P, Sauguet, L. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
8SH3
 
 | | Pendrin in complex with iodide | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, IODIDE ION, ... | | Authors: | Wang, L, Hoang, A, Zhou, M. | | Deposit date: | 2023-04-13 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
8SWK
 
 | | Cryo-EM structure of NLRP3 closed hexamer | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
6G41
 
 | | Crystal structure of SeMet-labeled mavirus penton protein | | Descriptor: | Minor capsid protein | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8VKT
 
 | | Crystallographic structure of dimetalated DapE from Enterococcus faecium | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Probable succinyl-diaminopimelate desuccinylase, ... | | Authors: | Gonzalez-Segura, L, Diaz-Vilchis, A, Terrazas-Lopez, M, Diaz-Sanchez, A.G. | | Deposit date: | 2024-01-09 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | The three-dimensional structure of DapE from Enterococcus faecium reveals new insights into DapE/ArgE subfamily ligand specificity.
Int.J.Biol.Macromol., 270, 2024
|
|
4AYG
 
 | | Lactobacillus reuteri N-terminally truncated glucansucrase GTF180 in orthorhombic apo-form | | Descriptor: | ACETIC ACID, CALCIUM ION, GLUCANSUCRASE, ... | | Authors: | Pijning, T, Vujicic-Zagar, A, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2012-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility of Truncated and Full-Length Glucansucrase Gtf180 Enzymes from Lactobacillus Reuteri 180.
FEBS J., 281, 2014
|
|
8CGC
 
 | | Structure of CSF1R in complex with a pyrollopyrimidine (compound 23) | | Descriptor: | (2S)-2-hydroxybutanedioic acid, GLYCEROL, Macrophage colony-stimulating factor 1 receptor, ... | | Authors: | Aarhus, T.I, Bjornstad, F, Wolowczyk, C, Larsen, K.U, Rognstad, L, Leithaug, T, Unger, A, Habenberger, P, Wolff, A, Bjorkoy, G, Pridans, C, Eickhoff, J, Klebl, B, Hoff, B.H, Sundby, E. | | Deposit date: | 2023-02-03 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Synthesis and Development of Highly Selective Pyrrolo[2,3- d ]pyrimidine CSF1R Inhibitors Targeting the Autoinhibited Form.
J.Med.Chem., 66, 2023
|
|
5Z2X
 
 | | Structure of Alcohol dehydrogenase from Kluyveromyces polyspora(KpADH) | | Descriptor: | 1,2-ETHANEDIOL, Alcohol dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, Y, Zhou, J.Y, Hou, X.D, Xu, G.C, Wu, L, Rao, Y.J, ZHou, J.H, Ni, Y. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
1OB7
 
 | | Cephaibol C | | Descriptor: | CEPHAIBOL C, ETHANOL, SODIUM ION | | Authors: | Bunkoczi, G, Schiell, M, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2003-01-24 | | Release date: | 2003-12-11 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Crystal Structures of Cephaibols
J.Pept.Sci., 9, 2003
|
|
8SWE
 
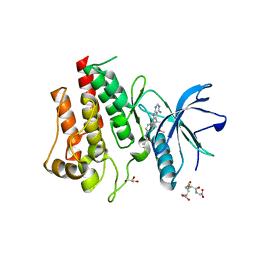 | | FGFR2 Kinase Domain Bound to Reversible Inhibitor Cmpd 3 | | Descriptor: | Fibroblast growth factor receptor 2, GLUTATHIONE, GLYCEROL, ... | | Authors: | Valverde, R, Foster, L. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of lirafugratinib (RLY-4008), a highly selective irreversible small-molecule inhibitor of FGFR2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SGW
 
 | | Pendrin in complex with chloride | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Wang, L, Hoang, A, Zhou, M. | | Deposit date: | 2023-04-13 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
1F8V
 
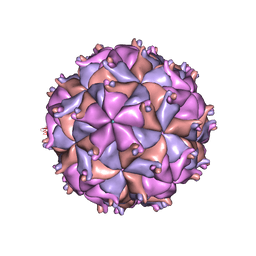 | | THE STRUCTURE OF PARIACOTO VIRUS REVEALS A DODECAHEDRAL CAGE OF DUPLEX RNA | | Descriptor: | CALCIUM ION, MATURE CAPSID PROTEIN BETA, MATURE CAPSID PROTEIN GAMMA, ... | | Authors: | Tang, L, Johnson, K.N, Ball, L.A, Lin, T, Yeager, M, Johnson, J.E. | | Deposit date: | 2000-07-05 | | Release date: | 2000-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of pariacoto virus reveals a dodecahedral cage of duplex RNA.
Nat.Struct.Biol., 8, 2001
|
|
8SHC
 
 | | Pendrin in complex with Niflumic acid | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}NICOTINIC ACID, CHLORIDE ION, ... | | Authors: | Wang, L, Hoang, A, Zhou, M. | | Deposit date: | 2023-04-13 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
8SXN
 
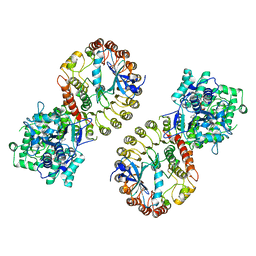 | | Structure of NLRP3 and NEK7 complex | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-22 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
4BEC
 
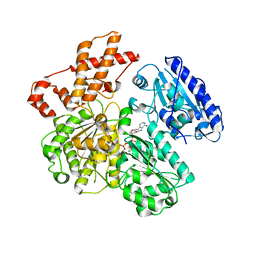 | | MUTANT (K220A) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, TYPE I RESTRICTION ENZYME HSDR | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
8SWF
 
 | | Cryo-EM structure of NLRP3 open octamer | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
