8BQY
 
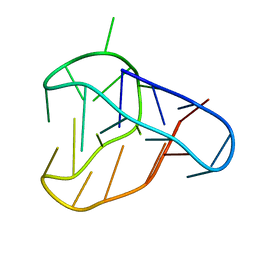 | | An i-motif domain able to undergo pH-dependent conformational transitions (acidic structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*(DNR)P*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*(DNR)P*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-11-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
8R60
 
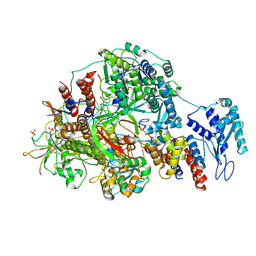 | | 1918 H1N1 Viral polymerase heterotrimer in complex with 4 repeat serine-5 phosphorylated PolII peptide | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2023-11-19 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural and functional characterization of the interaction between the influenza A virus RNA polymerase and the CTD of host RNA polymerase II.
J.Virol., 98, 2024
|
|
1F2U
 
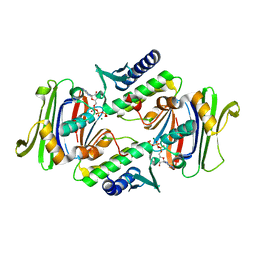 | | Crystal Structure of RAD50 ABC-ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RAD50 ABC-ATPASE | | Authors: | Hopfner, K.P, Karcher, A, Shin, D.S, Craig, L. | | Deposit date: | 2000-05-29 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily.
Cell(Cambridge,Mass.), 101, 2000
|
|
8RQF
 
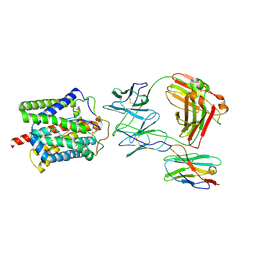 | | Cryo-EM structure of human NTCP-Bulevirtide complex | | Descriptor: | Fab-specific nanobody, Heavy chain of Fab3, Light chain of Fab3, ... | | Authors: | Liu, H, Zakrzewicz, D, Nosol, K, Irobalieva, R.N, Mukherjee, S, Bang-Soerensen, R, Goldmann, N, Kunz, S, Rossi, L, Kossiakoff, A.A, Urban, S, Glebe, D, Geyer, J, Locher, K.P. | | Deposit date: | 2024-01-18 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure of antiviral drug bulevirtide bound to hepatitis B and D virus receptor protein NTCP.
Nat Commun, 15, 2024
|
|
3SY0
 
 | | S25-2- A(2-8)-A(2-4)KDO trisaccharide complex | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, MAGNESIUM ION, S25-2 FAB (IGG1K) heavy chain, ... | | Authors: | Nguyen, H.P, Seto, N.O, Mackenzie, C.R, Brade, L, Kosma, P, Brade, H, Evans, S.V. | | Deposit date: | 2011-07-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Germline antibody recognition of distinct carbohydrate epitopes
NAT.STRUCT.BIOL., 10, 2003
|
|
1P56
 
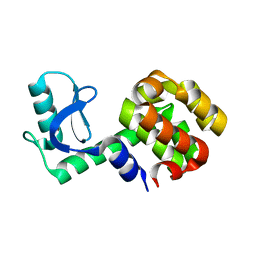 | |
1F46
 
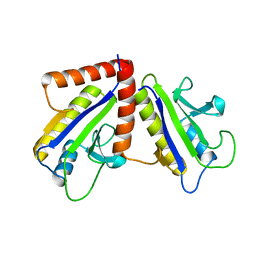 | | THE BACTERIAL CELL-DIVISION PROTEIN ZIPA AND ITS INTERACTION WITH AN FTSZ FRAGMENT REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | CELL DIVISION PROTEIN ZIPA | | Authors: | Mosyak, L, Zhang, Y, Glasfeld, E, Stahl, M, Somers, W.S. | | Deposit date: | 2000-06-07 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography.
EMBO J., 19, 2000
|
|
4C05
 
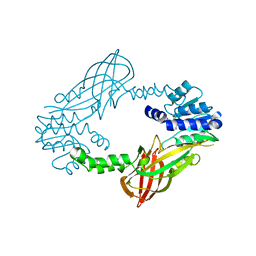 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with SAH | | Descriptor: | PROTEIN ARGININE N-METHYLTRANSFERASE 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
5YHG
 
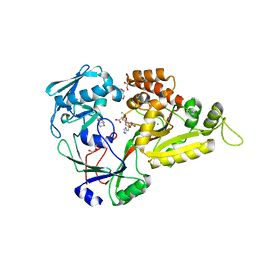 | | The crystal structure of Staphylococcus aureus CntA in complex with staphylopine and zinc | | Descriptor: | (2~{S})-4-[[(2~{R})-3-(1~{H}-imidazol-4-yl)-1-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-2-[[(2~{S})-1-oxidanyl-1-oxidanylidene-propan-2-yl]amino]butanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ji, Q, Song, L. | | Deposit date: | 2017-09-28 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mechanistic insights into staphylopine-mediated metal acquisition
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4C1I
 
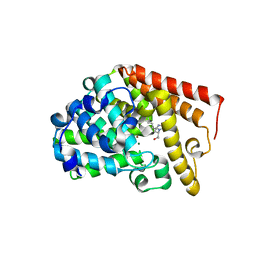 | | Selective Inhibitors of PDE2, PDE9, and PDE10: Modulators of Activity of the Central Nervous System | | Descriptor: | (2S,3R)-3-(6-amino-9H-purin-9-yl)nonan-2-ol, CGMP-DEPENDENT 3', 5'-CYCLIC PHOSPHODIESTERASE, ... | | Authors: | Jorgensen, M, Kehler, J, Langgard, M, Svenstrup, N, Tagmose, L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chapter 4: Selective Inhibitors of Pde2, Pde9, and Pde10: Modulators of Activity of the Central Nervous System
To be Published
|
|
1OZZ
 
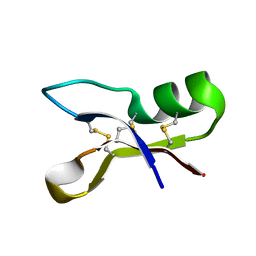 | | NMR structure of antifungal defensin ARD1 from Archaeoprepona demophon | | Descriptor: | defensin ARD1 | | Authors: | Landon, C, Guenneugues, M, Barbault, F, Legrain, M, Menin, L, Schott, V, Vovelle, F, Dimarcq, J.L. | | Deposit date: | 2003-04-10 | | Release date: | 2004-03-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Lead optimization of antifungal peptides with 3D NMR structures analysis.
Protein Sci., 13, 2004
|
|
4BTM
 
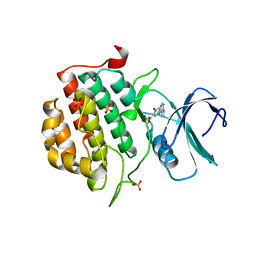 | | TTBK1 in complex with inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Xue, Y, Wan, P, Hillertz, P, Schweikart, F, Zhao, Y, Wissler, L, Dekker, N. | | Deposit date: | 2013-06-18 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | X-Ray Structural Analysis of Tau-Tubulin Kinase 1 and its Interactions with Small Molecular Inhibitors.
Chemmedchem, 8, 2013
|
|
1EZL
 
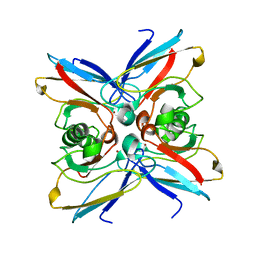 | | CRYSTAL STRUCTURE OF THE DISULPHIDE BOND-DEFICIENT AZURIN MUTANT C3A/C26A: HOW IMPORTANT IS THE S-S BOND FOR FOLDING AND STABILITY? | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Bonander, N, Leckner, J, Guo, H, Karlsson, B.G, Sjolin, L. | | Deposit date: | 2000-05-11 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the disulfide bond-deficient azurin mutant C3A/C26A: how important is the S-S bond for folding and stability?
Eur.J.Biochem., 267, 2000
|
|
1P75
 
 | | Crystal structure of EHV4-TK complexed with TP5A | | Descriptor: | P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, SULFATE ION, Thymidine kinase | | Authors: | Gardberg, A, Shuvalova, L, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2003-04-30 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis for the dual thymidine and thymidylate kinase activity of herpes thymidine kinases.
Structure, 11, 2003
|
|
5MWU
 
 | | Crystal structure of the periplasmic nickel-binding protein NikA from Escherichia coli in complex with Ru(bpza)(CO)2Cl | | Descriptor: | ACETATE ION, CARBON MONOXIDE, CHLORIDE ION, ... | | Authors: | Cavazza, C, Lopez, S, Rondot, L, Iannello, M, Boeri-Erba, E, Burzlaff, N, Strinitz, F, Jorge-Robin, A, Marchi-Delapierre, C, Menage, S. | | Deposit date: | 2017-01-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efficient conversion of alkenes to chlorohydrins by a Ru-based artificial enzyme
To Be Published
|
|
1P7R
 
 | | CRYSTAL STRUCTURE OF REDUCED, CO-EXPOSED COMPLEX OF CYTOCHROME P450CAM WITH (S)-(-)-NICOTINE | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, Cytochrome P450-cam, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Strickler, M, Goldstein, B.M, Maxfield, K, Shireman, L, Kim, G, Matteson, D, Jones, J.P. | | Deposit date: | 2003-05-05 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystallographic Studies on the Complex Behavior of Nicotine Binding to P450cam (CYP101)(dagger).
Biochemistry, 42, 2003
|
|
4C4W
 
 | | Structure of a rare, non-standard sequence k-turn bound by L7Ae protein | | Descriptor: | 50S RIBOSOMAL PROTEIN L7AE, DIHYDROGENPHOSPHATE ION, TSKT-23, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2013-09-09 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of a Rare, Non-Standard Sequence K-Turn Bound by L7Ae Protein
Nucleic Acids Res., 42, 2014
|
|
4C6R
 
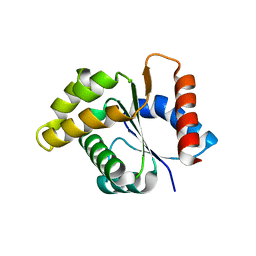 | | Crystal structure of the TIR domain from the Arabidopsis Thaliana disease resistance protein RPS4 | | Descriptor: | DISEASE RESISTANCE PROTEIN RPS4 | | Authors: | Williams, S.J, Sohn, K.H, Wan, L, Bernoux, M, Ma, Y, Segonzac, C, Ve, T, Sarris, P, Ericsson, D.J, Saucet, S.B, Zhang, X, Parker, J, Dodds, P.N, Jones, J.D.G, Kobe, B. | | Deposit date: | 2013-09-19 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Assembly and Function of a Heterodimeric Plant Immune Receptor.
Science, 344, 2014
|
|
8UD0
 
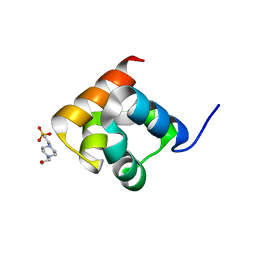 | |
1P8L
 
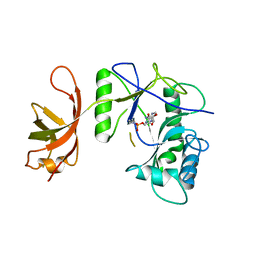 | | New Crystal Structure of Chlorella Virus DNA Ligase-Adenylate | | Descriptor: | ADENOSINE MONOPHOSPHATE, PBCV-1 DNA ligase | | Authors: | Odell, M, Malinina, L, Teplova, M, Shuman, S. | | Deposit date: | 2003-05-07 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Analysis of the DNA Joining Repertoire of Chlorella Virus DNA ligase and a New Crystal Structure of the Ligase-Adenylate Intermediate
Nucleic Acids Res., 31, 2003
|
|
8RB8
 
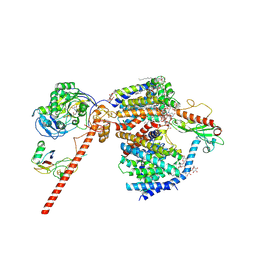 | | Cryo-EM structure of the NADH:ferredoxin oxidoreductase RNF from Azotobacter vinelandii, purified with 2-ME/TCEP, NADH added | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhang, L, Einsle, O. | | Deposit date: | 2023-12-03 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Architecture of the RNF1 complex that drives biological nitrogen fixation.
Nat.Chem.Biol., 20, 2024
|
|
5MVG
 
 | | Crystal structure of non-amyloidogenic light chain dimer M7 | | Descriptor: | GLYCEROL, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-16 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
1P00
 
 | | NMR structure of ETD151, mutant of the antifungal defensin ARD1 from Archaeoprepona demophon | | Descriptor: | defensin ARD1 | | Authors: | Landon, C, Guenneugues, M, Barbault, F, Legrain, M, Menin, L, Schott, V, Vovelle, F, Dimarcq, J.L. | | Deposit date: | 2003-04-10 | | Release date: | 2004-03-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Lead optimization of antifungal peptides with 3D NMR structures analysis.
Protein Sci., 13, 2004
|
|
5XZG
 
 | | Mouse cGAS bound to the inhibitor RU521 | | Descriptor: | 2-(4,5-dichloro-1H-benzimidazol-2-yl)-5-methyl-4-[(1R)-3-oxo-1,3-dihydro-2-benzofuran-1-yl]-1,2-dihydro-3H-pyrazol-3-one, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*G)-3'), ... | | Authors: | Vincent, J, Adura, C, Gao, P, Luz, A, Lama, L, Asano, Y, Okamoto, R, Imaeda, T, Aida, J, Rothamel, K, Gogakos, T, Steinberg, J, Reasoner, S, Aso, K, Tuschl, T, Patel, D.J, Glickman, J.F, Ascano, M. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Nat Commun, 8, 2017
|
|
6F6D
 
 | | The catalytic domain of KDM6B in complex with H3(17-33)K18IA21M peptide | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Histone 3 peptide H3(17-33)K18IA21M, ... | | Authors: | Jones, S.E, Olsen, L, Gajhede, M. | | Deposit date: | 2017-12-05 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81810677 Å) | | Cite: | Structural Basis of Histone Demethylase KDM6B Histone 3 Lysine 27 Specificity.
Biochemistry, 57, 2018
|
|
