5DGF
 
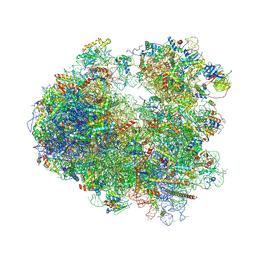 | | Complex of yeast 80S ribosome with hypusine-containing/non-modified eIF5A and/or a peptidyl-tRNA analog | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | Deposit date: | 2015-08-27 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome
To Be Published
|
|
6DDZ
 
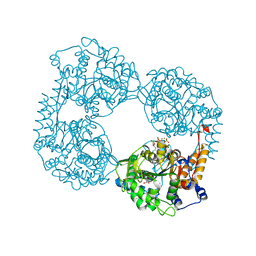 | | Crystal structure of the double mutant (D52N/R238W) of NT5C2-537X in the active state, Northeast Structural Genomics Target | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, GLYCEROL, ... | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
1ZMZ
 
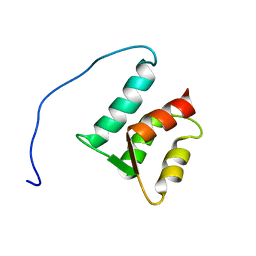 | | Solution structure of the N-terminal domain (M1-S98) of human centrin 2 | | Descriptor: | Centrin-2 | | Authors: | Yang, A, Miron, S, Duchambon, P, Assairi, L, Blouquit, Y, Craescu, C.T. | | Deposit date: | 2005-05-11 | | Release date: | 2006-04-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The N-terminal domain of human centrin 2 has a closed structure, binds calcium with a very low affinity, and plays a role in the protein self-assembly
Biochemistry, 45, 2006
|
|
8Q7J
 
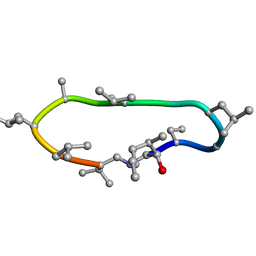 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability | | Descriptor: | CYCLOSPORIN A | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
1RIO
 
 | | Structure of bacteriophage lambda cI-NTD in complex with sigma-region4 of Thermus aquaticus bound to DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 27-MER, CALCIUM ION, ... | | Authors: | Jain, D, Nickels, B.E, Sun, L, Hochschild, A, Darst, S.A. | | Deposit date: | 2003-11-17 | | Release date: | 2004-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a ternary transcription activation complex.
Mol.Cell, 13, 2004
|
|
8QAS
 
 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. NMR structure of Omphalotin A in methanol / water indoleOut conformation. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
1YYL
 
 | | crystal structure of CD4M33, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 gp120 envelope glycoprotein and anti-HIV-1 antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD4M33, scorpion-toxin mimic of CD4, ... | | Authors: | Huang, C.C, Stricher, F, Martin, L, Decker, J.M, Majeed, S, Barthe, P, Hendrickson, W.A, Robinson, J, Roumestand, C, Sodroski, J, Wyatt, R, Shaw, G.M, Vita, C, Kwong, P.D. | | Deposit date: | 2005-02-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth.
Structure, 13, 2005
|
|
1RGA
 
 | |
8QF2
 
 | | Beta-L-Arabinofurano-cyclitol Aziridines are Cysteine-directed Broad-spectrum Inhibitors and Activity-based Probes for Retaining Beta-L-arabinofuranosidases | | Descriptor: | (1~{S},2~{S},3~{S},4~{R})-4-azanyl-3-(hydroxymethyl)cyclopentane-1,2-diol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Borlandelli, V, Offen, W.A, Moroz, O, Nin-Hill, A, McGregor, N, Binkhorst, L, Armstrong, Z, Ishiwata, A, Artola, M, Rovira, C, Davies, G.J, Overkleeft, H. | | Deposit date: | 2023-09-02 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | beta-l- Arabino furano-cyclitol Aziridines Are Covalent Broad-Spectrum Inhibitors and Activity-Based Probes for Retaining beta-l-Arabinofuranosidases.
Acs Chem.Biol., 18, 2023
|
|
8QBP
 
 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. NMR structure of Omphalotin A in methanol / water indoleOut conformation. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
8QF8
 
 | | GH146 beta-L-arabinofuranosidase from Bacteroides thetaioatomicron in complex with beta-l-arabinofurano cyclophellitol aziridine | | Descriptor: | (1~{S},2~{S},3~{S},4~{R})-4-azanyl-3-(hydroxymethyl)cyclopentane-1,2-diol, (1~{S},2~{S},3~{S},4~{S},5~{S})-4-(hydroxymethyl)-6-azabicyclo[3.1.0]hexane-2,3-diol, Glycosyl hydrolase, ... | | Authors: | Borlandelli, V, Offen, W, Moroz, O.V, Nin-Hill, A, McGregor, N, Binkhorst, L, Armstrong, Z, Ishiwata, A, Artola, M, Rovira, C, Davies, G.J, Overkleeft, H. | | Deposit date: | 2023-09-04 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | beta-l- Arabino furano-cyclitol Aziridines Are Covalent Broad-Spectrum Inhibitors and Activity-Based Probes for Retaining beta-l-Arabinofuranosidases.
Acs Chem.Biol., 18, 2023
|
|
1RKJ
 
 | | Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target | | Descriptor: | 5'-R(*GP*GP*AP*UP*GP*CP*CP*UP*CP*CP*CP*GP*AP*GP*UP*GP*CP*AP*UP*CP*C)-3', Nucleolin | | Authors: | Johansson, C, Finger, L.D, Trantirek, L, Mueller, T.D, Kim, S, Laird-Offringa, I.A, Feigon, J. | | Deposit date: | 2003-11-21 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target.
J.Mol.Biol., 337, 2004
|
|
8Q6S
 
 | | A carbohydrate esterase family 15 (CE15) glucuronoyl esterase from Phocaeicola vulgatus ATCC 8482 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Putative acetyl xylan esterase, ... | | Authors: | Mazurkewich, S, Seveso, A, Banerjee, S, Lo Leggio, L, Larsbrink, J. | | Deposit date: | 2023-08-14 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Polysaccharide utilization loci from Bacteroidota encode CE15 enzymes with possible roles in cleaving pectin-lignin bonds.
Appl.Environ.Microbiol., 90, 2024
|
|
5CSF
 
 | | S100B-RSK1 crystal structure A | | Descriptor: | CALCIUM ION, Protein S100-B, Ribosomal protein S6 kinase alpha-1 | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Ribosomal S6 Kinase 1 (RSK1) Inhibition by S100B Protein: MODULATION OF THE EXTRACELLULAR SIGNAL-REGULATED KINASE (ERK) SIGNALING CASCADE IN A CALCIUM-DEPENDENT WAY.
J.Biol.Chem., 291, 2016
|
|
1YWN
 
 | | Vegfr2 in complex with a novel 4-amino-furo[2,3-d]pyrimidine | | Descriptor: | N-{4-[4-AMINO-6-(4-METHOXYPHENYL)FURO[2,3-D]PYRIMIDIN-5-YL]PHENYL}-N'-[2-FLUORO-5-(TRIFLUOROMETHYL)PHENYL]UREA, Vascular endothelial growth factor receptor 2 | | Authors: | Miyazaki, Y, Matsunaga, S, Tang, J, Maeda, Y, Nakano, M, Philippe, R.J, Shibahara, M, Liu, W, Sato, H, Wang, L, Nolte, R.T. | | Deposit date: | 2005-02-18 | | Release date: | 2005-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Novel 4-amino-furo[2,3-d]pyrimidines as Tie-2 and VEGFR2 dual inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1YX0
 
 | | Solution Structure of Bacillus subtilis Protein ysnE: The Northeast Structural Genomics Consortium Target SR220 | | Descriptor: | hypothetical protein ysnE | | Authors: | Liu, G, Ma, L, Shen, Y, Xiao, R, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Bacillus subtilis Protein ysnE
To be Published
|
|
8QEF
 
 | | A carbohydrate esterase family 15 (CE15) glucuronoyl esterase from Phocaeicola ATCC 8482 bound to novel ligand. | | Descriptor: | 1,2-ETHANEDIOL, Putative acetyl xylan esterase, beta-D-galactopyranuronic acid | | Authors: | Banerjee, S, Poulsen, J.N, Mazurkewich, S, Seveso, A, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2023-08-31 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Polysaccharide utilization loci from Bacteroidota encode CE15 enzymes with possible roles in cleaving pectin-lignin bonds.
Appl.Environ.Microbiol., 90, 2024
|
|
8QCL
 
 | | A carbohydrate esterase family 15 (CE15) glucuronoyl esterase from Phocaeicola vulgatus ATCC 8482 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Banerjee, S, Poulsen, J.N, Mazurkewich, S, Seveso, A, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2023-08-27 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Polysaccharide utilization loci from Bacteroidota encode CE15 enzymes with possible roles in cleaving pectin-lignin bonds.
Appl.Environ.Microbiol., 90, 2024
|
|
1RKN
 
 | | Solution structure of 1-110 fragment of Staphylococcal Nuclease with G88W mutation | | Descriptor: | Thermonuclease | | Authors: | Liu, D.S, Feng, Y.G, Ye, K.Q, Shan, L, Wang, J.F. | | Deposit date: | 2003-11-22 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
8R05
 
 | | Photorhabdus lamondii ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.5 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1YZ6
 
 | |
5ER4
 
 | | Crystal Structure of Calcium-loaded S100B bound to SC0025 | | Descriptor: | 6-methyl-5,6,6~{a},7-tetrahydro-4~{H}-dibenzo[de,g]quinoline-10,11-diol, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Melville, Z.E, Aligholizadeh, E, Fang, L, Alasady, M.J, Pierce, A.D, Wilder, P.T, MacKerell Jr, A.D, Weber, D.J. | | Deposit date: | 2015-11-13 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Novel protein-inhibitor interactions in site 3 of Ca(2+)-bound S100B as discovered by X-ray crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8R03
 
 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.0 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1Z05
 
 | | Crystal structure of the ROK family transcriptional regulator, homolog of E.coli MLC protein. | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, SULFATE ION, ... | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-01 | | Release date: | 2005-03-08 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the ROK family transcriptional regulator, homolog of E.coli MLC protein.
To be Published
|
|
1Z94
 
 | | X-Ray Crystal Structure of Protein CV1439 from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CvR12. | | Descriptor: | conserved hypothetical protein | | Authors: | Kuzin, A, Vorobiev, S.M, Yong, W, Forouhar, F, Xio, R, Ma, L.-C, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-03-31 | | Release date: | 2005-05-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Hypothetical Protein CV1439 from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CVR12.
To be Published
|
|
