1E8J
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS ZINC RUBREDOXIN, NMR, 20 STRUCTURES | | Descriptor: | RUBREDOXIN | | Authors: | Lamosa, P, Brennan, L, Vis, H, Turner, D.L, Santos, H. | | Deposit date: | 2000-09-21 | | Release date: | 2001-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Desulfovibrio gigas rubredoxin: a model for studying protein stabilization by compatible solutes.
Extremophiles, 5, 2001
|
|
1PPG
 
 | | The refined 2.3 angstroms crystal structure of human leukocyte elastase in a complex with a valine chloromethyl ketone inhibitor | | Descriptor: | HUMAN LEUKOCYTE ELASTASE, MEO-SUCCINYL-ALA-ALA-PRO-VAL CHLOROMETHYLKETONE, alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bode, W, Wei, A-Z. | | Deposit date: | 1991-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The refined 2.3 A crystal structure of human leukocyte elastase in a complex with a valine chloromethyl ketone inhibitor.
FEBS Lett., 234, 1988
|
|
1GLQ
 
 | |
4CA4
 
 | | Crystal structure of FimH lectin domain with the Tyr48Ala mutation, in complex with heptyl alpha-D-mannopyrannoside | | Descriptor: | FIMH, heptyl alpha-D-mannopyranoside | | Authors: | Rabbani, S, Bouckaert, J, Zalewski, A, Preston, R, Eid, S, Thompson, A, Puorger, C, Glockshuber, R, Ernst, B. | | Deposit date: | 2013-10-06 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Mutation of Tyr137 of the universal Escherichia coli fimbrial adhesin FimH relaxes the tyrosine gate prior to mannose binding.
IUCrJ, 4, 2017
|
|
6X84
 
 | |
1DCD
 
 | | DESULFOREDOXIN COMPLEXED WITH CD2+ | | Descriptor: | CADMIUM ION, PROTEIN (DESULFOREDOXIN) | | Authors: | Archer, M, Carvalho, A.L, Teixeira, S, Romao, M.J. | | Deposit date: | 1999-03-20 | | Release date: | 1999-07-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies by X-ray diffraction on metal substituted desulforedoxin, a rubredoxin-type protein.
Protein Sci., 8, 1999
|
|
1FJS
 
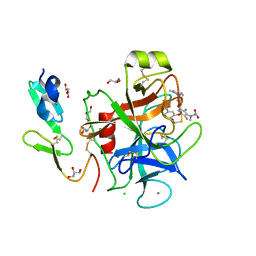 | | CRYSTAL STRUCTURE OF THE INHIBITOR ZK-807834 (CI-1031) COMPLEXED WITH FACTOR XA | | Descriptor: | CALCIUM ION, CHLORIDE ION, COAGULATION FACTOR XA, ... | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2000-08-08 | | Release date: | 2000-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Preparation, characterization, and the crystal structure of the inhibitor ZK-807834 (CI-1031) complexed with factor Xa.
Biochemistry, 39, 2000
|
|
1G6X
 
 | | ULTRA HIGH RESOLUTION STRUCTURE OF BOVINE PANCREATIC TRYPSIN INHIBITOR (BPTI) MUTANT WITH ALTERED BINDING LOOP SEQUENCE | | Descriptor: | 1,2-ETHANEDIOL, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Addlagatta, A, Czapinska, H, Krzywda, S, Otlewski, J, Jaskolski, M. | | Deposit date: | 2000-11-08 | | Release date: | 2001-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Ultrahigh-resolution structure of a BPTI mutant.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G65
 
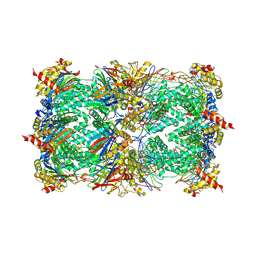 | | Crystal structure of epoxomicin:20s proteasome reveals a molecular basis for selectivity of alpha,beta-epoxyketone proteasome inhibitors | | Descriptor: | EPOXOMICIN (peptide inhibitor), MAGNESIUM ION, Proteasome component C1, ... | | Authors: | Groll, M, Kim, K.B, Kairies, N, Huber, R, Crews, C. | | Deposit date: | 2000-11-03 | | Release date: | 2000-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Epoxomicin:20S Proteasome reveals a molecular basis for selectivity of alpha,beta-Epoxyketone Proteasome Inhibitors
J.Am.Chem.Soc., 122, 2000
|
|
1G41
 
 | | CRYSTAL STRUCTURE OF HSLU HAEMOPHILUS INFLUENZAE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK PROTEIN HSLU, SULFATE ION | | Authors: | Trame, C.B, McKay, D.B. | | Deposit date: | 2000-10-25 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Haemophilus influenzae HslU protein in crystals with one-dimensional disorder twinning.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1DE7
 
 | |
1E5Y
 
 | |
5OH0
 
 | | The Cryo-Electron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Hospenthal, M.K, Costa, T.R.D, Redzej, A, Waksman, G. | | Deposit date: | 2017-07-13 | | Release date: | 2017-11-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The Cryoelectron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod.
Structure, 25, 2017
|
|
1CTI
 
 | |
1CE5
 
 | | BOVINE PANCREAS BETA-TRYPSIN IN COMPLEX WITH BENZAMIDINE | | Descriptor: | BENZAMIDINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ota, N, Stroupe, C, Ferreira-Da-Silva, J.M.S, Shah, S.S, Mares-Guia, M, Brunger, A.T. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Non-Boltzmann thermodynamic integration (NBTI) for macromolecular systems: relative free energy of binding of trypsin to benzamidine and benzylamine.
Proteins, 37, 1999
|
|
1ETS
 
 | |
1FC2
 
 | |
3BCI
 
 | |
1E5Z
 
 | |
1ETR
 
 | | REFINED 2.3 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF BOVINE THROMBIN COMPLEXES FORMED WITH THE BENZAMIDINE AND ARGININE-BASED THROMBIN INHIBITORS NAPAP, 4-TAPAP AND MQPA: A STARTING POINT FOR IMPROVING ANTITHROMBOTICS | | Descriptor: | EPSILON-THROMBIN, amino{[(4S)-5-[(2R,4R)-2-carboxy-4-methylpiperidin-1-yl]-4-({[(3R)-3-methyl-1,2,3,4-tetrahydroquinolin-8-yl]sulfonyl}amino)-5-oxopentyl]amino}methaniminium | | Authors: | Bode, W, Brandstetter, H. | | Deposit date: | 1992-07-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined 2.3 A X-ray crystal structure of bovine thrombin complexes formed with the benzamidine and arginine-based thrombin inhibitors NAPAP, 4-TAPAP and MQPA. A starting point for improving antithrombotics.
J.Mol.Biol., 226, 1992
|
|
1G4B
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
1ETT
 
 | |
1G4A
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
3BCK
 
 | |
3F4R
 
 | | Crystal structure of Wolbachia pipientis alpha-DsbA1 | | Descriptor: | PENTAETHYLENE GLYCOL, Putative uncharacterized protein, TRIETHYLENE GLYCOL | | Authors: | Kurz, M, Heras, B, Martin, J.L. | | Deposit date: | 2008-11-02 | | Release date: | 2009-03-24 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of the Oxidoreductase alpha-DsbA1 from Wolbachia pipientis.
ANTIOXID.REDOX SIGNAL., 11, 2009
|
|
