1SWX
 
 | | Crystal structure of a human glycolipid transfer protein in apo-form | | Descriptor: | Glycolipid transfer protein, HEXANE | | Authors: | Malinina, L, Malakhova, M.L, Teplov, A, Brown, R.E, Patel, D.J. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for glycosphingolipid transfer specificity.
Nature, 430, 2004
|
|
1SX6
 
 | | Crystal structure of human Glycolipid Transfer protein in lactosylceramide-bound form | | Descriptor: | Glycolipid transfer protein, N-OCTANE, OLEIC ACID, ... | | Authors: | Malinina, L, Malakhova, M.L, Teplov, A, Brown, R.E, Patel, D.J. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for glycosphingolipid transfer specificity.
Nature, 430, 2004
|
|
2LW1
 
 | | The C-terminal domain of the Uup protein is a DNA-binding coiled coil motif | | Descriptor: | ABC transporter ATP-binding protein uup | | Authors: | Carlier, L, Haase, A.S, Burgos Zepeda, M.Y, Dassa, E, Lequin, O. | | Deposit date: | 2012-07-19 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-terminal domain of the Uup protein is a DNA-binding coiled coil motif.
J.Struct.Biol., 180, 2012
|
|
5DF1
 
 | | Iridoid synthase from Catharanthus roseus - ternary complex with NADP+ and geranic acid | | Descriptor: | (2E)-3,7-dimethylocta-2,6-dienoic acid, 1,2-ETHANEDIOL, IMIDAZOLE, ... | | Authors: | Caputi, L, Kries, H, Stevenson, C.E.M, Kamileen, M.O, Sherden, N.H, Geu-Flores, F, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural determinants of reductive terpene cyclization in iridoid biosynthesis.
Nat.Chem.Biol., 12, 2016
|
|
1SXR
 
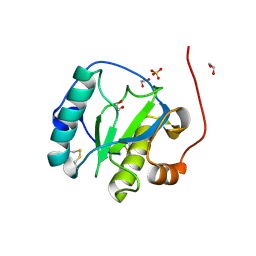 | | Drosophila Peptidoglycan Recognition Protein (PGRP)-SA | | Descriptor: | 1,2-ETHANEDIOL, Peptidoglycan recognition protein SA CG11709-PA, SULFATE ION | | Authors: | Reiser, J.B, Teyton, L, Wilson, I.A. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of the Drosophila peptidoglycan recognition protein (PGRP)-SA at 1.56 A resolution
J.Mol.Biol., 340, 2004
|
|
8ON5
 
 | |
1XK0
 
 | | Crystal Structures of the G139A, G139A-NO and G143H Mutants of Human Heme Oxygenase-1 | | Descriptor: | Heme oxygenase 1, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lad, L, Koshkin, A, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2004-09-26 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structures of the G139A, G139A-NO and G143H mutants of human heme oxygenase-1. A finely tuned hydrogen-bonding network controls oxygenase versus peroxidase activity.
J.Biol.Inorg.Chem., 10, 2005
|
|
1XJB
 
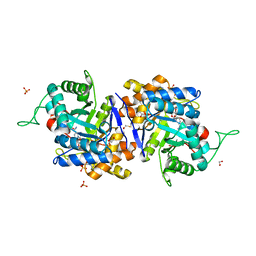 | | Crystal structure of human type 3 3alpha-hydroxysteroid dehydrogenase in complex with NADP(H), citrate and acetate molecules | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aldo-keto reductase family 1 member C2, ... | | Authors: | Couture, J.-F, Pereira de Jesus-Tran, K, Roy, A.-M, Legrand, P, Cantin, L, Cote, P.-L, Luu-The, V, Labrie, F, Breton, R. | | Deposit date: | 2004-09-23 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of crystal structures of human type 3 3alpha-hydroxysteroid dehydrogenase reveals an "induced-fit" mechanism and a conserved basic motif involved in the binding of androgen
Protein Sci., 14, 2005
|
|
5DIR
 
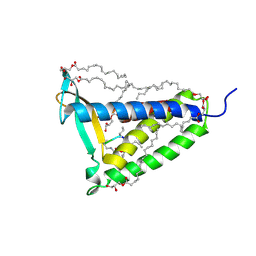 | | membrane protein at 2.8 Angstroms | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Globomycin, Lipoprotein signal peptidase | | Authors: | Vogeley, L, El Arnaout, T, Bailey, J, Boland, C, Caffrey, M. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of lipoprotein signal peptidase II action and inhibition by the antibiotic globomycin.
Science, 351, 2016
|
|
5DJ1
 
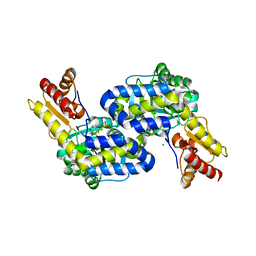 | | Structure of the PLP-Dependent L-Arginine Hydroxylase MppP Holoenzyme | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Silvaggi, N.R, Han, L. | | Deposit date: | 2015-09-01 | | Release date: | 2015-11-25 | | Last modified: | 2017-05-03 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Streptomyces wadayamensis MppP Is a Pyridoxal 5'-Phosphate-Dependent l-Arginine alpha-Deaminase, gamma-Hydroxylase in the Enduracididine Biosynthetic Pathway.
Biochemistry, 54, 2015
|
|
6D0N
 
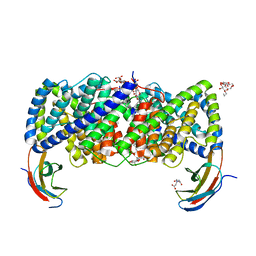 | | Crystal structure of a CLC-type fluoride/proton antiporter, V319G mutant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CLC-type fluoride/proton antiporter, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Last, N.B, Stockbridge, R.B, Wilson, A.E, Shane, T, Kolmakova-Partensky, L, Koide, A, Koide, S, Miller, C. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A CLC-type F-/H+antiporter in ion-swapped conformations.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5DQR
 
 | |
1XLS
 
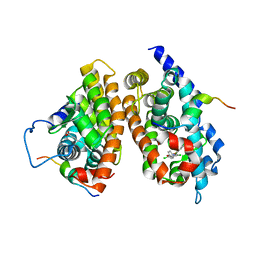 | | Crystal structure of the mouse CAR/RXR LBD heterodimer bound to TCPOBOP and 9cRA and a TIF2 peptide containg the third LXXLL motifs | | Descriptor: | (9cis)-retinoic acid, 3,5-DICHLORO-2-{4-[(3,5-DICHLOROPYRIDIN-2-YL)OXY]PHENOXY}PYRIDINE, Nuclear receptor coactivator 2, ... | | Authors: | Suino, K, peng, L, Reynolds, R, Li, Y, Cha, J.-Y, Repa, J.J, Kliewer, S.A, Xu, H.E. | | Deposit date: | 2004-09-30 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization.
Mol.Cell, 16, 2004
|
|
6CG6
 
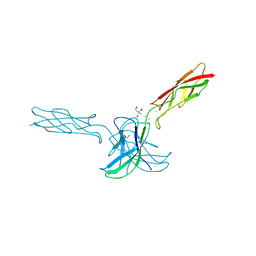 | | mouse cadherin-10 EC1-2 adhesive fragment | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cadherin-10, ... | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2018-02-19 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior.
Cell Rep, 23, 2018
|
|
1THZ
 
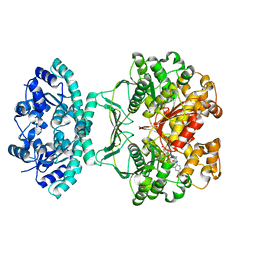 | | Crystal Structure of Avian AICAR Transformylase in Complex with a Novel Inhibitor Identified by Virtual Ligand Screening | | Descriptor: | 2-{(E)-[5-HYDROXY-3-METHYL-1-(2-METHYL-4-SULFOPHENYL)-1H-PYRAZOL-4-YL]DIAZENYL}-4-SULFOBENZOIC ACID, Bifunctional purine biosynthesis protein PURH, POTASSIUM ION | | Authors: | Xu, L, Li, C, Olson, A.J, Wilson, I.A. | | Deposit date: | 2004-06-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of avian aminoimidazole-4-carboxamide ribonucleotide transformylase in complex with a novel non-folate inhibitor identified by virtual ligand screening.
J.Biol.Chem., 279, 2004
|
|
1XPJ
 
 | | Crystal Structure of MCSG Target APC26283 from Vibrio cholerae | | Descriptor: | L(+)-TARTARIC ACID, MERCURY (II) ION, hypothetical protein | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-08 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of MCSG Target APC26283 from Vibrio cholerae
To be Published
|
|
1T2L
 
 | | Three Crystal Structures of Human Coactosin-like Protein | | Descriptor: | Coactosin-like protein | | Authors: | Liu, L, Wei, Z, Chen, Z, Wang, Y, Gong, W. | | Deposit date: | 2004-04-22 | | Release date: | 2004-11-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Coactosin-like Protein
J.Mol.Biol., 344, 2004
|
|
6JXD
 
 | |
6CGU
 
 | | mouse cadherin-6 EC1-2 adhesive fragment | | Descriptor: | CALCIUM ION, Cadherin-6 | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior.
Cell Rep, 23, 2018
|
|
8OI2
 
 | | Crystal structure of Alb1 megabody in complex with human serum albumin | | Descriptor: | Alb1 Megabody, Albumin | | Authors: | De Felice, S, Zoia, G, Romanyuk, Z, Pardon, E, Steyaert, J, Angelini, A, Cendron, L. | | Deposit date: | 2023-03-22 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of human serum albumin in complex with megabody reveals unique human and murine cross-reactive binding site.
Protein Sci., 33, 2024
|
|
5DLL
 
 | | Aminopeptidase N (pepN) from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Aminopeptidase N, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Borek, D, Raczynska, J, Dubrovska, I, Grimshaw, S, Minasov, G, Shuvalova, L, Kwon, K, Anderson, W.F, Otwinowski, Z, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Aminopeptidase N (pepN) from Francisella tularensis subsp. tularensis SCHU S4
To Be Published
|
|
8ORH
 
 | | Knockout of GMC-oxidoreductase genes reveals that functional redundancy preserves mimivirus essential functions | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Alempic, J.M, Bisio, H, Villalta, A, Santini, S, Lartigue, A, Schmitt, A, Bugnot, C, Notaro, A, Belmudes, L, Adrait, A, Poirot, O, Ptchelkine, D, De Castro, C, Coute, Y, Abergel, C. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Functional redundancy revealed by the deletion of the mimivirus GMC-oxidoreductase genes.
Microlife, 5, 2024
|
|
6JZE
 
 | | Crystal structure of VASH2-SVBP complex with the magic triangle I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 2 | | Authors: | Chen, Z, Ling, Y, Zeyuan, G, Zhu, L. | | Deposit date: | 2019-05-01 | | Release date: | 2019-08-07 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis of tubulin detyrosination by VASH2/SVBP heterodimer.
Nat Commun, 10, 2019
|
|
1U6L
 
 | |
5DNA
 
 | | Crystal structure of Candida boidinii formate dehydrogenase | | Descriptor: | FORMATE DEHYDROGENASE, SULFATE ION | | Authors: | Guo, Q, Gakhar, L, Wichersham, K, Francis, K, Vardi-Kilshtain, A, Major, D.T, Cheatum, C.M, Kohen, A. | | Deposit date: | 2015-09-09 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Kinetic Studies of Formate Dehydrogenase from Candida boidinii.
Biochemistry, 55, 2016
|
|
