9ATI
 
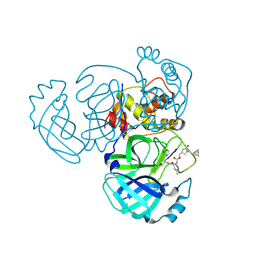 | | Crystal structure of MERS 3CL protease in complex with a racemic bicyclo[2.2.1]heptenyl-methyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-{[N-({[(3S)-1-{[(1R,2R,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(3S)-1-{[(1R,2R,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ATH
 
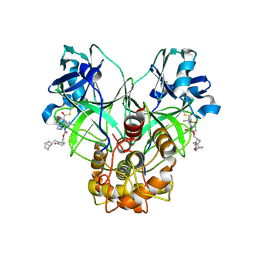 | | Crystal structure of MERS 3CL protease in complex with a methylbicyclo[2.2.1]heptene 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-{[N-({[(2S)-1-{[(1R,2S,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-{[(1R,2S,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9AT7
 
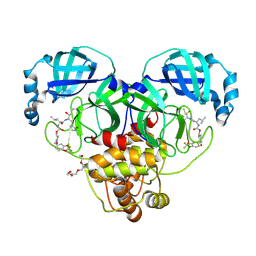 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a 2,2-difluoro-5-methylbenzo[1,3]dioxole 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(2,2-difluoro-2H-1,3-benzodioxol-5-yl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(2,2-difluoro-2H-1,3-benzodioxol-5-yl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ATS
 
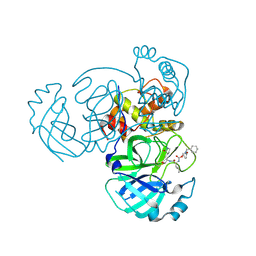 | | Crystal structure of MERS 3CL protease in complex with a methylcyclohexyl 2-pyrrolidone inhibitor (S-enantiomer) | | Descriptor: | (1S,2S)-2-{[N-({[(2S)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-27 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9AT6
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a methylbicyclo[2.2.1]heptene 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-{[N-({[(2S)-1-{[(1R,2S,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-{[(1R,2S,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ATJ
 
 | | Crystal structure of MERS 3CL protease in complex with a m-chlorobenzyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(3-chlorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(3-chlorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ASZ
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a phenylethyl 2-pyrrolidone inhibitor | | Descriptor: | (1S,2S)-1-hydroxy-2-{[N-({[(2S)-5-oxo-1-(2-phenylethyl)pyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ATD
 
 | | Crystal structure of MERS 3CL protease in complex with a ethylcyclohexyl 2-pyrrolidone inhibitor (S-enantiomer) inhibitor | | Descriptor: | (1R,2S)-2-{[N-({[(2S)-1-(2-cyclohexylethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-(2-cyclohexylethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9AT1
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a methylcyclohexyl 2-pyrrolidone inhibitor (R-enantiomer) | | Descriptor: | (1S,2S)-2-{[N-({[(2R)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ATF
 
 | | Crystal structure of MERS 3CL protease in complex with a 1-methyl-4,4-difluorocyclohexyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(4,4-difluorocyclohexyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(4,4-difluorocyclohexyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
2PL2
 
 | | Crystal structure of TTC0263: a thermophilic TPR protein in Thermus thermophilus HB27 | | Descriptor: | Hypothetical conserved protein TTC0263 | | Authors: | Lim, H, Kim, K, Han, D, Oh, J. | | Deposit date: | 2007-04-18 | | Release date: | 2008-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of TTC0263, a thermophilic TPR protein from Thermus thermophilus HB27.
Mol.Cell, 24, 2007
|
|
8ECZ
 
 | | Bovine Fab 4C1 | | Descriptor: | 4C1 Fab heavy chain, 4C1 Fab light chain, PHOSPHATE ION | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EDF
 
 | |
8ECV
 
 | | Bovine Fab 2F12 | | Descriptor: | 2F12 Fab Heavy chain, 2F12 Fab Light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ED1
 
 | | Bovine Fab 5C1 | | Descriptor: | 5C1 Fab heavy chain, 5C1 Fab light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ECQ
 
 | | Bovine Fab 2G3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2G3 Fab Heavy chain, 2G3 Fab Light chain, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8V6U
 
 | | 5HT2AR-miniGq heterotrimer in complex with a novel agonist obtained from large scale docking | | Descriptor: | 4-{(3R)-1-[(1R)-1-(pyrimidin-2-yl)ethyl]piperidin-3-yl}phenol, 5-hydroxytryptamine receptor 2A, G protein alpha-subunit q (Gi2-mini-Gq chimera), ... | | Authors: | Gumpper, R.H, Wang, L, Kapolka, N, Skiniotis, G, Roth, B.L. | | Deposit date: | 2023-12-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | AlphaFold2 structures guide prospective ligand discovery.
Science, 384, 2024
|
|
8GSV
 
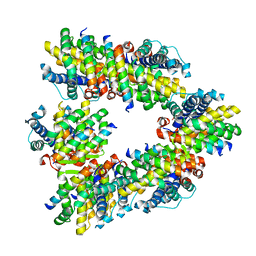 | | Crystal structure of human BAK in complex with the Pxt1 BH3 domain | | Descriptor: | Bcl-2 homologous antagonist/killer, Peroxisomal testis-specific protein 1 | | Authors: | Lim, D, Ku, B. | | Deposit date: | 2022-09-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for proapoptotic activation of Bak by the noncanonical BH3-only protein Pxt1.
Plos Biol., 21, 2023
|
|
2Q7F
 
 | |
3LQY
 
 | | Crystal structure of putative isochorismatase hydrolase from Oleispira antarctica | | Descriptor: | GLYCEROL, putative isochorismatase hydrolase | | Authors: | Goral, A, Chruszcz, M, Kagan, O, Cymborowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-10 | | Release date: | 2010-03-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative isochorismatase hydrolase from Oleispira antarctica.
J.Struct.Funct.Genom., 13, 2012
|
|
5Y41
 
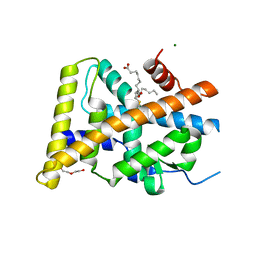 | | Crystal Structure of LIGAND-BOUND NURR1-LBD | | Descriptor: | (13E,15S)-15-hydroxy-9-oxoprosta-10,13-dien-1-oic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sreekanth, R, Lescar, J, Yoon, H.S. | | Deposit date: | 2017-07-31 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | PGE1 and PGA1 bind to Nurr1 and activate its transcriptional function.
Nat.Chem.Biol., 2020
|
|
6ISS
 
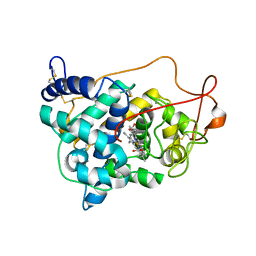 | | Lignin peroxidase H8 triple mutant S49C/A67C/H239 | | Descriptor: | CALCIUM ION, Ligninase H8, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Seo, H, Son, H, Kim, K.-J. | | Deposit date: | 2018-11-19 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Extra disulfide and ionic salt bridge improves the thermostability of lignin peroxidase H8 under acidic condition
Enzyme.Microb.Technol., 148, 2021
|
|
4WKY
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmN KS2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-ketoacyl synthase, GLYCEROL, ... | | Authors: | Cuff, M.E, Mack, J.C, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2FI7
 
 | |
5Z9G
 
 | | Crystal structure of KAI2 | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
