1SGZ
 
 | |
5CMV
 
 | | Ultrafast dynamics in myoglobin: dark-state, CO-ligated structure | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
5CND
 
 | | Ultrafast dynamics in myoglobin: 3 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
8JT8
 
 | | Crystal structure of 5-HT2AR in complex with (R)-IHCH-7179 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Chen, Z, Fan, L, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 187, 2024
|
|
1SQS
 
 | | X-Ray Crystal Structure Protein SP1951 of Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR27. | | Descriptor: | L(+)-TARTARIC ACID, conserved hypothetical protein | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-19 | | Release date: | 2004-03-30 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM, 8, 2007
|
|
6IPU
 
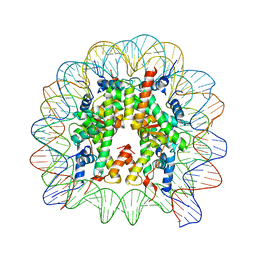 | |
6ANO
 
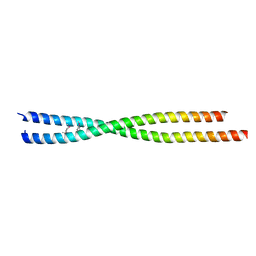 | | Crystal structure of human FLASH N-terminal domain | | Descriptor: | CASP8-associated protein 2 | | Authors: | Aik, W.S, Tong, L. | | Deposit date: | 2017-08-14 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The N-terminal domains of FLASH and Lsm11 form a 2:1 heterotrimer for histone pre-mRNA 3'-end processing.
PLoS ONE, 12, 2017
|
|
1SR3
 
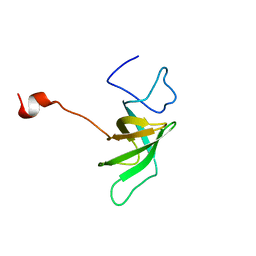 | |
5C15
 
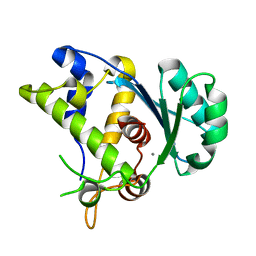 | |
1SH9
 
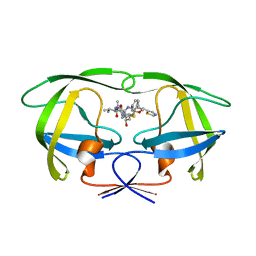 | | Comparing the Accumulation of Active Site and Non-active Site Mutations in the HIV-1 Protease | | Descriptor: | POL polyprotein, RITONAVIR | | Authors: | Clemente, J.C, Moose, R.E, Hemrajani, R, Govindasamy, L, Reutzel, R, McKenna, R, Abanje-McKenna, M, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2004-02-25 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparing the Accumulation of Active- and Nonactive-Site Mutations in the HIV-1 Protease.
Biochemistry, 43, 2004
|
|
6AU0
 
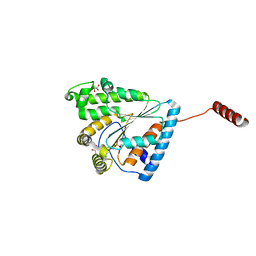 | | Crystal structure of PPK2 (Class III) in complex with bisphosphonate inhibitor (2-((3,5-dichlorophenyl)amino)ethane-1,1-diyl)diphosphonic acid | | Descriptor: | GLYCEROL, Polyphosphate:AMP phosphotransferase, {[(3,5-dichlorophenyl)amino]methylene}bis(phosphonic acid) | | Authors: | Nocek, B, Ruszkowski, M, Joachimiak, A, Berlicki, L, Yakunin, A. | | Deposit date: | 2017-08-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into Substrate Selectivity and Activity of Bacterial Polyphosphate Kinases
Acs Catalysis, 8, 2018
|
|
1SGW
 
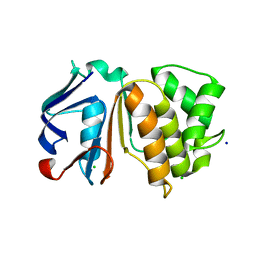 | | Putative ABC transporter (ATP-binding protein) from Pyrococcus furiosus Pfu-867808-001 | | Descriptor: | CHLORIDE ION, SODIUM ION, putative ABC transporter | | Authors: | Liu, Z.J, Tempel, W, Shah, A, Chen, L, Lee, D, Kelley, L.-L.C, Dillard, B.D, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-02-24 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Putative ABC transporter (ATP-binding protein) from Pyrococcus furiosus Pfu-867808-001
To be Published
|
|
6AWI
 
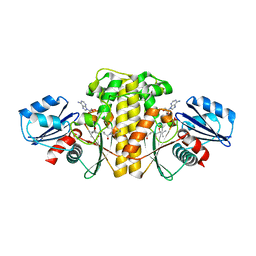 | | Staphylococcus aureus Type II pantothenate kinase in complex with ADP and pantothenate analog Deoxy-N5Pan | | Descriptor: | (2R)-2-hydroxy-3,3-dimethyl-N-[3-oxo-3-(pentylamino)propyl]butanamide, ADENOSINE-5'-DIPHOSPHATE, Type II pantothenate kinase | | Authors: | Chen, Y, Antoshchenko, T, Strauss, E, Barnard, L, Huang, Y.H. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based identification of uncompetitive inhibitors for Staphylococcus aureus pantothenate kinase.
To Be Published
|
|
1SP3
 
 | | Crystal structure of octaheme cytochrome c from Shewanella oneidensis | | Descriptor: | HEME C, THIOCYANATE ION, cytochrome c, ... | | Authors: | Mowat, C.G, Rothery, E, Miles, C.S, McIver, L, Doherty, M.K, Drewette, K, Taylor, P, Walkinshaw, M.D, Chapman, S.K, Reid, G.A. | | Deposit date: | 2004-03-16 | | Release date: | 2004-09-21 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Octaheme tetrathionate reductase is a respiratory enzyme with novel heme ligation.
Nat.Struct.Mol.Biol., 11, 2004
|
|
6AXA
 
 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with (3S)-5-fluoro-3-hydroxy-1,3-dihydroindol-2-one | | Descriptor: | (3S)-5-fluoro-3-hydroxy-1,3-dihydro-2H-indol-2-one, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-09-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 5-fluoro-3-hydroxy-1,3-dihydroindol-2-one
To Be Published
|
|
6AZQ
 
 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | CALCIUM ION, Putative amidase, dicarbonimidic diamide | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
5C7O
 
 | | Structure of human testis-specific glyceraldehyde-3-phosphate dehydrogenase holo form with NAD+ | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Betts, L, Machius, M, Danshina, P, O'Brien, D. | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Structural analyses to identify selective inhibitors of glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme.
Mol. Hum. Reprod., 22, 2016
|
|
1WQC
 
 | | An unusual fold for potassium channel blockers : NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis | | Descriptor: | OmTx1 | | Authors: | Chagot, B, Pimentel, C, Dai, L, Pil, J, Tytgat, J, Nakajima, T, Corzo, G, Darbon, H, Ferrat, G. | | Deposit date: | 2004-09-27 | | Release date: | 2005-01-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis
Biochem.J., 388, 2005
|
|
1SMK
 
 | | Mature and translocatable forms of glyoxysomal malate dehydrogenase have different activities and stabilities but similar crystal structures | | Descriptor: | CITRIC ACID, Malate dehydrogenase, glyoxysomal | | Authors: | Cox, B, Chit, M.M, Weaver, T, Bailey, J, Gietl, C, Bell, E, Banaszak, L. | | Deposit date: | 2004-03-09 | | Release date: | 2005-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Organelle and translocatable forms of glyoxysomal malate dehydrogenase. The effect of the N-terminal presequence.
Febs J., 272, 2005
|
|
1SMO
 
 | | Crystal Structure of Human Triggering Receptor Expressed on Myeloid Cells 1 (TREM-1) at 1.47 . | | Descriptor: | L(+)-TARTARIC ACID, triggering receptor expressed on myeloid cells 1 | | Authors: | Kelker, M.S, Foss, T.R, Peti, W, Teyton, L, Kelly, J.W, Wilson, I.A. | | Deposit date: | 2004-03-09 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structure of Human Triggering Receptor Expressed on Myeloid Cells 1 (TREM-1) at 1.47A.
J.Mol.Biol., 342, 2004
|
|
5CB2
 
 | | the structure of candida albicans Sey1p in complex with GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein SEY1 | | Authors: | Yan, L, Sun, S, Wang, W, Shi, J, Hu, X, Wang, S, Rao, Z, Hu, J, Lou, Z. | | Deposit date: | 2015-06-30 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the yeast dynamin-like GTPase Sey1p provide insight into homotypic ER fusion
J.Cell Biol., 210, 2015
|
|
6B37
 
 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 1,3-benzothiazol-2-ol | | Descriptor: | 1,3-benzothiazol-2-ol, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-09-21 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 1,3-benzothiazol-2-ol
To Be Published
|
|
6B3L
 
 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 2,3-dihydro-1H-inden-2-amine | | Descriptor: | DIMETHYL SULFOXIDE, INDAN-2-AMINE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-09-22 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 2,3-dihydro-1H-inden-2-amine
To Be Published
|
|
1WSU
 
 | | C-terminal domain of elongation factor selB complexed with SECIS RNA | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*UP*GP*CP*CP*GP*GP*UP*CP*U*GP*GP*CP*AP*AP*CP*GP*CP*C)-3', Selenocysteine-specific elongation factor | | Authors: | Yoshizawa, S, Rasubala, L, Ose, T, Kohda, D, Fourmy, D, Maenaka, K. | | Deposit date: | 2004-11-11 | | Release date: | 2005-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for mRNA recognition by elongation factor SelB
Nat.Struct.Mol.Biol., 12, 2005
|
|
1SP9
 
 | | 4-Hydroxyphenylpyruvate Dioxygenase | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, FE (II) ION | | Authors: | Fritze, I.M, Linden, L, Freigang, J, Auerbach, G, Huber, R, Steinbacher, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structures of Zea mays and Arabidopsis 4-Hydroxyphenylpyruvate Dioxygenase
Plant Physiol., 134, 2004
|
|
