7YTB
 
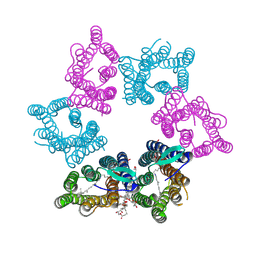 | | Crystal structure of Kin4B8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Kin4B8, RETINAL | | Authors: | Murakoshi, S, Chazan, A, Shihoya, W, Beja, O, Nureki, O. | | Deposit date: | 2022-08-14 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phototrophy by antenna-containing rhodopsin pumps in aquatic environments.
Nature, 615, 2023
|
|
9J5V
 
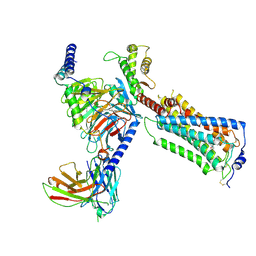 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to CpY | | Descriptor: | CpY, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2024-08-13 | | Release date: | 2024-11-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural mechanisms of potent lysophosphatidic acid receptor 1 activation by nonlipid basic agonists.
Commun Biol, 7, 2024
|
|
9IZR
 
 | | Cryo-EM structure of CasLambda2-crRNA-target DNA ternary complex in the NTS-cleaving state | | Descriptor: | 2-DEOXY-ADENOSINE -5'-THIO-MONOPHOSPHATE, 2-DEOXY-CYTIDINE-5'-THIOPHOSPHORATE, Cas Lambda2, ... | | Authors: | Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-08-01 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for target DNA cleavage and guide RNA processing by CRISPR-Cas lambda 2.
Commun Biol, 8, 2025
|
|
9IZS
 
 | | Cryo-EM structure of CasLambda2-crRNA-target DNA ternary complex in the TS-cleaving state | | Descriptor: | Cas Lambda2, DNA (40-MER), MAGNESIUM ION, ... | | Authors: | Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-08-01 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis for target DNA cleavage and guide RNA processing by CRISPR-Cas lambda 2.
Commun Biol, 8, 2025
|
|
9J7W
 
 | | Channel Rhodospin from Klebsormidium nitens (KnChR) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, KnChR, RETINAL | | Authors: | Wang, Y.Z, Akasaka, H, Tanaka, T, Sano, F.K, Shihoya, W, Nureki, O. | | Deposit date: | 2024-08-20 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Cryo-EM structure of a blue-shifted channelrhodopsin from Klebsormidium nitens.
Nat Commun, 16, 2025
|
|
9IZP
 
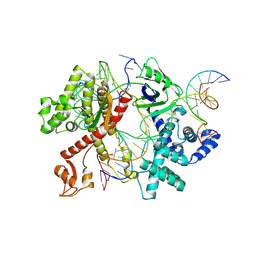 | | Cryo-EM structure of CasLambda2-crRNA-target DNA ternary complex in the incompetent state | | Descriptor: | Cas Lambda2, DNA (40-MER), MAGNESIUM ION, ... | | Authors: | Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-08-01 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural basis for target DNA cleavage and guide RNA processing by CRISPR-Cas lambda 2.
Commun Biol, 8, 2025
|
|
9IZM
 
 | | Cryo-EM structure of CasLambda2-crRNA binary complex | | Descriptor: | Cas Lambda2, MAGNESIUM ION, RNA (58-MER) | | Authors: | Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-08-01 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis for target DNA cleavage and guide RNA processing by CRISPR-Cas lambda 2.
Commun Biol, 8, 2025
|
|
9IZQ
 
 | | Cryo-EM structure of CasLambda2-crRNA-target DNA ternary complex in the intermediate state | | Descriptor: | Cas Lambda2, DNA (40-MER), MAGNESIUM ION, ... | | Authors: | Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-08-01 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for target DNA cleavage and guide RNA processing by CRISPR-Cas lambda 2.
Commun Biol, 8, 2025
|
|
1A8H
 
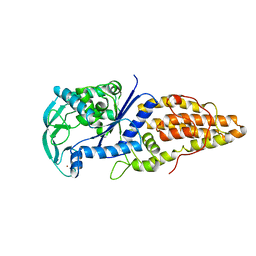 | | METHIONYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS | | Descriptor: | METHIONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sugiura, I, Nureki, O, Ugaji, Y, Kuwabara, S, Lober, B, Giege, R, Moras, D, Yokoyama, S, Konno, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules.
Structure, 8, 2000
|
|
6IU9
 
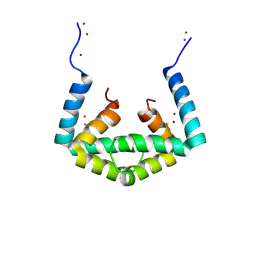 | | Crystal structure of cytoplasmic metal binding domain with iron ions | | Descriptor: | FE (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IGK
 
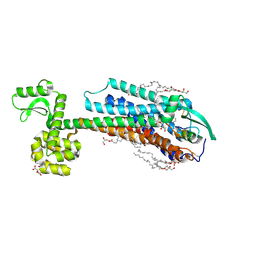 | | Crystal Structure of human ETB receptor in complex with Endothelin-3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRIC ACID, Endothelin receptor type B,Endolysin,Endothelin receptor type B, ... | | Authors: | Shihoya, W, Izume, T, Inoue, A, Yamashita, K, Kadji, F.M.N, Hirata, K, Aoki, J, Nishizawa, T, Nureki, O. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human ETBreceptor provide mechanistic insight into receptor activation and partial activation.
Nat Commun, 9, 2018
|
|
6IRZ
 
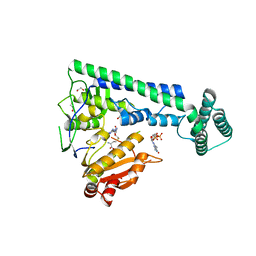 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | Descriptor: | 1,2-ETHANEDIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, PDX1 C-terminal-inhibiting factor 1, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IJF
 
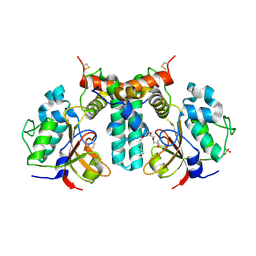 | | Crystal structure of the type VI effector-immunity complex (Tae4-Tai4) from Agrobacterium tumefaciens | | Descriptor: | PENTAETHYLENE GLYCOL, SULFATE ION, Tae4, ... | | Authors: | Fukuhara, S, Nakane, T, Yamashita, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2018-10-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Agrobacterium tumefaciens type VI effector-immunity complex.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6IU5
 
 | | Crystal structure of cytoplasmic metal binding domain with zinc ions | | Descriptor: | CHLORIDE ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IRV
 
 | |
6IU4
 
 | | Crystal structure of iron transporter VIT1 with cobalt ion | | Descriptor: | COBALT (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Taniguchi, R, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IDS
 
 | | Crystal structure of Vibrio cholerae MATE transporter VcmN D35N mutant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
6IS0
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IU8
 
 | | Crystal structure of cytoplasmic metal binding domain with cobalt ions | | Descriptor: | COBALT (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IJE
 
 | | Crystal structure of the type VI amidase immunity (Tai4) from Agrobacterium tumefaciens | | Descriptor: | 1,2-ETHANEDIOL, Tai4 | | Authors: | Fukuhara, S, Nakane, T, Yamashita, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2018-10-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the Agrobacterium tumefaciens type VI effector-immunity complex.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6IGL
 
 | | Crystal Structure of human ETB receptor in complex with IRL1620 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRIC ACID, Endothelin receptor type B,Endolysin,Endothelin receptor type B, ... | | Authors: | Shihoya, W, Izume, T, Inoue, A, Yamashita, K, kadji, F.M.N, Hirata, K, Aoki, J, Nishizawa, T, Nureki, O. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of human ETBreceptor provide mechanistic insight into receptor activation and partial activation.
Nat Commun, 9, 2018
|
|
6IRW
 
 | | Crystal structure of the human cap-specific adenosine methyltransferase bound to SAH | | Descriptor: | Phosphorylated CTD-interacting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
4OO8
 
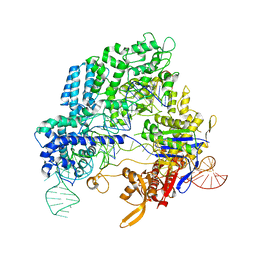 | | Crystal structure of Streptococcus pyogenes Cas9 in complex with guide RNA and target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*CP*CP*AP*GP*CP*CP*AP*AP*GP*CP*GP*CP*AP*CP*CP*TP*AP*AP*TP*TP*TP*CP*C)-3'), RNA (97-MER) | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2014-01-31 | | Release date: | 2014-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Cas9 in complex with guide RNA and target DNA
Cell(Cambridge,Mass.), 156, 2014
|
|
4P79
 
 | | Crystal structure of mouse claudin-15 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Claudin-15 | | Authors: | Suzuki, H, Nishizawa, T, Tani, K, Yamazaki, Y, Tamura, A, Ishitani, R, Dohmae, N, Tsukita, S, Nureki, O, Fujiyoshi, Y. | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a claudin provides insight into the architecture of tight junctions.
Science, 344, 2014
|
|
4PDN
 
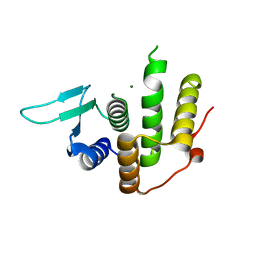 | | Crystal structure of E. coli YfcM | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Kobayashi, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2014-04-19 | | Release date: | 2015-03-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | The non-canonical hydroxylase structure of YfcM reveals a metal ion-coordination motif required for EF-P hydroxylation.
Nucleic Acids Res., 42, 2014
|
|
