2G3R
 
 | | Crystal Structure of 53BP1 tandem tudor domains at 1.2 A resolution | | Descriptor: | SULFATE ION, Tumor suppressor p53-binding protein 1 | | Authors: | Lee, J, Botuyan, M.V, Thompson, J.R, Mer, G. | | Deposit date: | 2006-02-20 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis for the Methylation State-Specific Recognition of Histone H4-K20 by 53BP1 and Crb2 in DNA Repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
2FHD
 
 | | Crystal structure of Crb2 tandem tudor domains | | Descriptor: | DNA repair protein rhp9/CRB2, PHOSPHATE ION | | Authors: | Lee, J, Botuyan, M.V, Thompson, J.R, Mer, G. | | Deposit date: | 2005-12-23 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
3Q66
 
 | |
3SZM
 
 | |
5UMR
 
 | | Crystal structure of N-terminal domain of human FACT complex subunit SSRP1 | | Descriptor: | FACT complex subunit SSRP1 | | Authors: | Su, D, Hu, Q, Thompson, J.R, Heroux, A, Botuyan, M.V, Mer, G. | | Deposit date: | 2017-01-29 | | Release date: | 2018-01-31 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of N-terminal domain of human FACT complex subunit SSRP1
To Be Published
|
|
8UPF
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168-UbcH5c | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-10-22 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
5UMS
 
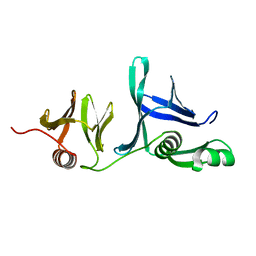 | |
3U3Z
 
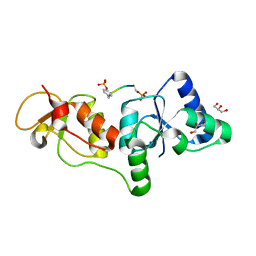 | | Structure of human microcephalin (MCPH1) tandem BRCT domains in complex with an H2A.X peptide phosphorylated at Ser139 and Tyr142 | | Descriptor: | GLYCEROL, Histone H2A.X peptide, Microcephalin | | Authors: | Singh, N, Thompson, J.R, Heroux, A, Mer, G. | | Deposit date: | 2011-10-06 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dual recognition of phosphoserine and phosphotyrosine in histone variant H2A.X by DNA damage response protein MCPH1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5UMU
 
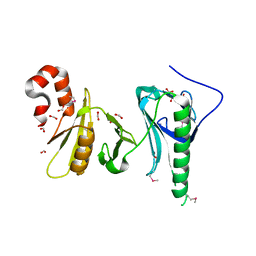 | | Crystal structure of the middle double PH domain of human FACT complex subunit SPT16 | | Descriptor: | ACETATE ION, FACT complex subunit SPT16, FORMIC ACID | | Authors: | Hu, Q, Thompson, J.R, Heroux, A, Su, D, Botuyan, M.V, Mer, G. | | Deposit date: | 2017-01-29 | | Release date: | 2018-01-31 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal structure of the middle double PH domain of human FACT complex subunit SPT16
To Be Published
|
|
5UMT
 
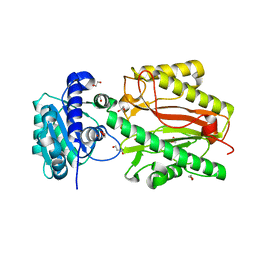 | | Crystal structure of N-terminal domain of human FACT complex subunit SPT16 | | Descriptor: | 1,2-ETHANEDIOL, FACT complex subunit SPT16, GLYCEROL | | Authors: | Su, D, Hu, Q, Thompson, J.R, Botuyan, M.V, Mer, G. | | Deposit date: | 2017-01-29 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Crystal structure of N-terminal domain of human FACT complex subunit SPT16
To Be Published
|
|
8TXV
 
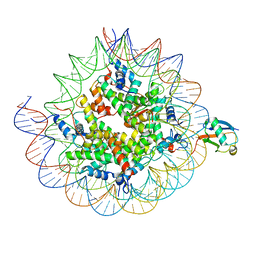 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 1) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TXX
 
 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 3) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TXW
 
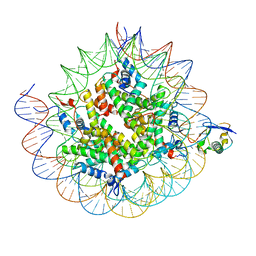 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 2) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
3Q68
 
 | |
5VWE
 
 | |
2QQR
 
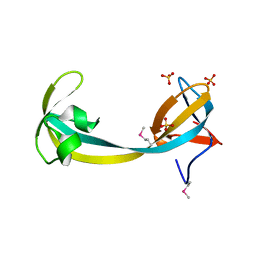 | | JMJD2A hybrid tudor domains | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, SULFATE ION | | Authors: | Lee, J, Botuyan, M.V, Mer, G. | | Deposit date: | 2007-07-26 | | Release date: | 2007-12-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3T1N
 
 | |
2QQS
 
 | |
3PA6
 
 | |
2LH0
 
 | |
2L0G
 
 | |
2L0F
 
 | |
2LVM
 
 | |
2K2W
 
 | | Second BRCT domain of NBS1 | | Descriptor: | Recombination and DNA repair protein | | Authors: | Xu, C, Cui, G, Botuyan, M, Mer, G. | | Deposit date: | 2008-04-14 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a second BRCT domain identified in the nijmegen breakage syndrome protein Nbs1 and its function in an MDC1-dependent localization of Nbs1 to DNA damage sites.
J.Mol.Biol., 381, 2008
|
|
2LY1
 
 | |
